Exploring Ribosome Code: Unveiling A Novel Dimension of Gene Regulation in Carcinogenesis
Yuen Gao1*
1Department of Physiology, Michigan State University, East Lansing, Michigan, USA
*Correspondence to: Yuen Gao, PhD, Assistant Professor, Department of Physiology, Michigan State University, 426 Auditorium Road, East Lansing, Michigan 48824, USA; Email: gaoyuen@msu.edu
Abstract
The central dogma of molecular biology has traditionally viewed ribosomes as uniform entities translating mRNA into proteins with mechanical precision. Recent findings challenge this perspective, revealing unexpected ribosome heterogeneity within cells, thus proposing a novel mechanism for diversifying protein synthesis. This concept parallels the “histone code” hypothesis, leading to the proposition of a “ribosome code” that suggests variations in ribosomal protein stoichiometry, paralogs, and modifications may preferentially translate certain mRNA populations. The role of ribosome heterogeneity in cancer pathogenesis, however, remains a subject of debate. Evidence suggests that alterations in ribosome composition, structure, and activity could influence abnormal protein synthesis, thus contributing to tumorigenesis. This is supported by the correlation between enhanced ribosome biogenesis, protein synthesis, and tumor growth, as well as the observed modifications in cancer ribosomes. Examining the nuances of ribosome heterogeneity, its influence on translational biases, and implications for tumor biology could significantly deepen our understanding of cancer’s molecular landscape, potentially unveiling novel therapeutic targets.
Keywords: ribosome code, cancer ribosome, gene regulation, carcinogenesis
Ever since its formulation, the central dogma of molecular biology has elucidated the route of genetic information flow within a biological system[1]. The conventional understanding has been that the ribosome, the cellular machinery responsible for translating mRNA into protein, carries out its function with almost mechanical precision. This view casts the ribosome as an essential, but largely passive, player in the production of effector proteins across the entire biological spectrum.
Recently, growing evidence challenges this long-held notion by demonstrating that ribosomes within a cell are far from homogenous[2,3]. Instead, a surprising level of ribosome heterogeneity exists within each cell. This unexpected diversity of ribosomes offers a novel mechanism to increase the variety of proteins produced in specific cells, tissues, and organisms. Similar with the concept of histone code that various post-translational modifications of histone proteins can influence the activation or suppression of transcription[4], ribosome code hypothesis postulates that ribosome with distinct stoichiometry, paralogs and modification of ribosomal proteins (RPs) and rRNA preferentially translating particular members of the message population over others[5].
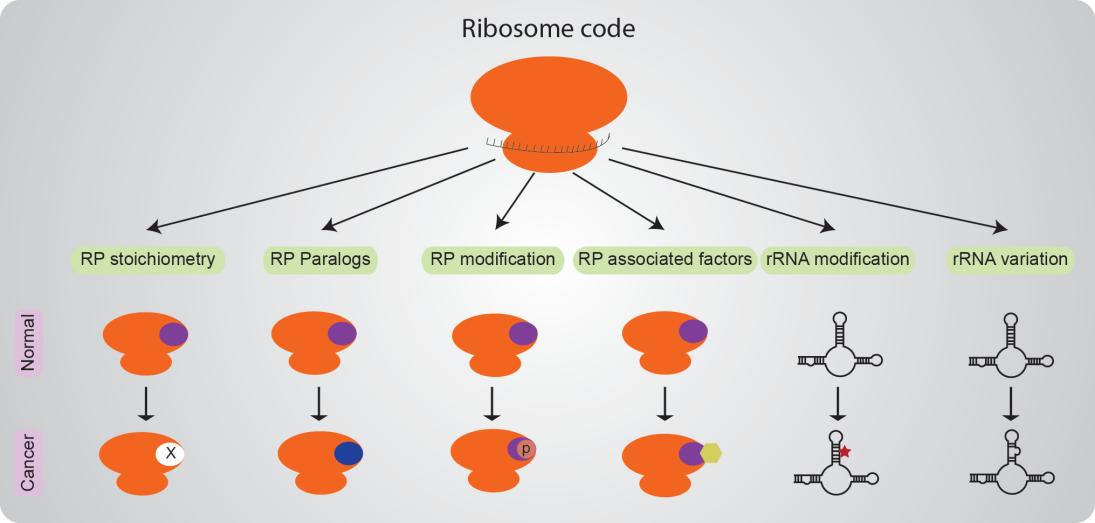
Meanwhile, whether cancer ribosomes exhibit alterations in their composition, structure, and activity and lead to abnormal protein synthesis and contribute to the development and progression of the carcinogenesis is still debating (Figure 1).
|
Figure 1. Ribosome code and cancer ribosome hypothesis.
Enhanced ribosome biogenesis and protein synthesis play a vital function in supporting the growth and proliferation of tumor cells[6,7]. Both the augmentation in ribosome quantities and modifications contribute to the development of tumors (Figure 1). An illustration of a recently evolved RP paralog with distinctive tissue-specific expression patterns is RPL39L, which exhibits increased expression in Hepatocellular carcinoma tumors[8]. In non-small cell lung cancer, the hyperphosphorylation of RP S6 serves as a predictive marker for unfavorable clinical survival[9]. Ribosome binding protein FMRP is suggested to play a role as a regulator that coordinates a complex barrier against antitumor immune responses[10]. To support the cancer ribosomes, Zhou et al.[11] reported that plasticity of rRNA 2’-O-methylation shifted protein translation toward a leukemia stem cell phenotype. Rothschild et al.[12] establish a curated atlas that connects the high expression of low abundant rRNA variants in cancer to tissue-specific biology, thereby linking ribosome variation to both cancer and tissue-specific functions. Collectively, cancer ribosomes are prevalent across various types of cancers.
By investigating the underpinnings of ribosome heterogeneity, the resulting translational biases, and their implications for tumor biology, we aim to foster a more comprehensive understanding of cancer’s molecular landscape and possibly unveil new targets for therapeutic interventions. We believe that addressing this topic could significantly enrich the ongoing discourse in cancer biology, and potentially inspire innovative strategies to counteract this devastating disease.
Acknowledgements
Not applicable.
Conflicts of Interest
The author declared no conflict of interest.
Author Contribution
Gao Y drafted and edited the manuscript, and approved the final version.
Abbreviation List
RP, Ribosomal protein
References
[1] Crick FH. On protein synthesis. Symp Soc Exp Biol, 1958; 12: 138-163.
[2] Gupta V, Warner JR. Ribosome-omics of the human ribosome. RNA, 2014; 20: 1004-1013. [DOI]
[3] Parks MM, Kurylo CM, Dass RA et al. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci Adv, 2018; 4: eaao0665. [DOI]
[4] Jenuwein T, Allis CD. Translating the histone code. Science, 2001; 293: 1074-1080. [DOI]
[5] Emmott E, Jovanovic M, Slavov N. Ribosome Stoichiometry: From Form to Function. Trends Biochem Sci, 2019; 44: 95-109. [DOI]
[6] Pelletier J, Thomas G, Volarević S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat Rev Cancer, 2018; 18: 51-63. [DOI]
[7] Bustelo XR, Dosil M. Ribosome biogenesis and cancer: Basic and translational challenges. Curr Opin Genet Dev, 2018; 48: 22-29. [DOI]
[8] Wong QW, Li J, Ng SR et al. RPL39L is an example of a recently evolved ribosomal protein paralog that shows highly specific tissue expression patterns and is upregulated in ESCs and HCC tumors. RNA Biol, 2014; 11: 33-41. [DOI]
[9] Chen BJ, Tan Z, Gao J et al. Hyperphosphorylation of ribosomal protein S6 predicts unfavorable clinical survival in non-small cell lung cancer. J Exp Clin Cancer Res, 2015; 34: 126. [DOI]
[10] Zeng Q, Saghafinia S, Chryplewicz A et al. Aberrant hyperexpression of the RNA binding protein FMRP in tumors mediates immune evasion. Science, 2022; 378: eabl7207. [DOI]
[11] Zhou F, Aroua N, Liu Y et al. A Dynamic rRNA Ribomethylome Drives Stemness in Acute Myeloid Leukemia. Cancer Discov, 2023; 13: 332-347. [DOI]
[12] Rothschild D, Susanto TT, Spence JP et al. A comprehensive rRNA variation atlas in health and disease. BioRxiv, 2023. [DOI]
Copyright © 2023 The Author(s). This open-access article is licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, sharing, adaptation, distribution, and reproduction in any medium, provided the original work is properly cited.




 Copyright ©
Copyright ©