Chemogenomics: Is A Promising Area for Drug Target and Discovery
Rakesh Devidas Amrutkar1*, Vaibhav G Bhamare2, Sachin N Kapse3
1Department of Pharmaceutical Chemistry, K. K. Wagh College of Pharmacy, Nasik, Maharashatra, India
2Department of Pharmaceutics, K. K. Wagh College of Pharmacy, Nasik, Maharashatra, India
3Department of Pharmaceutical Chemistry, Matoshri College of Pharmacy, Nasik, Maharashatra, India
*Correspondence to: Rakesh Devidas Amrutkar, PhD, Associate Professor, Department of Pharmaceutical Chemistry, K. K. Wagh College of Pharmacy, Hirabai Haridas Nagari, Mumbai-agra Road, Panchavati, Nashik 422003, Maharashatra, India; Email: rakesh_2504@yahoo.co.in
Abstract
The latest tools of genomics and chemistry and applies them to target and drug discovery is chemogenomoics. It is defined as the analysis of class of organic compound libraries against families of functionally related proteins called as receptor. chemogenomoics help to investigate biological target for the new drug entities. This deals with the systematic investigation of chemical-biological interactions. Study of chemogenomoics is the very important tool in medicinal chemistry for the development of new chemical entities) The main goal of chemogenomics is to identify the newer drugs and the targets because of number of chemical entities ware present so it is very difficult to identify the targets of said chemical compounds. The chemogenomic approach is the interaction of possible drug molecules with all probable targets In this regards we have reported significance and application of chemogenomoics.
Keywords: chemogenomoics, drug target, compounds libraries, drug discovery
1 INTRODUCTION
The term chemical biology, chemical genetics, and chemogenomoics are recent strategies in drug discovery and development. As per the literature serve the definitions of the said terms are somehow differ and conflicting. The term chemical biology means the study of biological systems, e.g., whole cells, under the influence of libraries of chemical compound. Chemical genetics is the devoted study of protein functions, e.g., signaling chains, considering of ligands which bind to certain proteins or interfere with protein-protein interaction; sometimes orthogonal ligand-protein pairs are generated to achieve selectivity for a certain protein. So the term chemogenomics principle based remove, the screening of the chemical universe, i.e., all possible chemical compounds, against the target universe, i.e., all proteins and other potential drug target[1]. Stuart Schreiber scientist at Harvard University and co-founder of the Broad Institute who reported the term chemical genetics in the year 1998 during the study of immunosuppressant therapy of Cyclosporina (Tacrolimus [FK506]) cyclo-philin interactions[1]. The findings of new chemical libraries of small molecules that are capable of interacting with any biological target through chemogenomoics is a novel way in drug discovery. (e.g., tyrosine kinase, PPARγ, etc.). The main goal of chemogenomics is to identify the newer drugs and the targets because of number of chemical entities ware present so it is very difficult to identify the targets of said chemical compounds. The chemogenomic approach is the interaction of possible drug molecules with all probable targets[2,3]. It has been reported that organic compounds targeting the products of 400-500 genes present in the human body, however, out of it only 120 of these genes are predicted in to the market as the targets of drugs[4,5].
Approximately 30,000-40,000 human genes[6,7] could be related with the disease and, a number of thousand human genes could be susceptible to drugs as per the Human genome project reports this drug-susceptibility and disease-associated genes will be probably allow the identification of genes that could serve as new drug targets for the development of new chemical entities[4]. According to the literature survey only around 500 different proteins are targeted by currently available drugs[8,9].
In this post-genomic world, the pharmaceutical industry rapidly discovers and develops medicines against new targets to improve the human health condition. Thepharmaceutical industries organized research and early development activities according to the therapeutic area the organizing their drug discovery efforts in this way, companies have sought to create efficiency by building a critical mass of expertise and experience in the biology of related diseases. Over the past 20-30 years, this organizational approach has proved successful for many companies. This strategy produces some synergies in early stage research and the greater efficiency in the later stages of clinical development and marketing of research and development resources along therapeutic area lines[10,11].
2 APPROACHES FOR CHEMOGENOMICS
The combination of effects of compounds on biological targets together with modern genomics technologies is nothing but chemogenomoics connect. There are two main experimental chemogenomoics methods are forward (classical) chemogenomoics and reverse chemogenomoics. Identification of drug targets by lead molecules which give a certain phenotype to cells or animals is called classical chemogenomoics. While specific interaction of protein (receptor) with validated phenotype by searching molecule interaction is called reverses chemogenomoics[12]. suitable collection of compounds is required for both of these approaches and an appropriate model system for screening the compounds and looking for the similar identification of biological targets and biologically active compounds. In the term of chemogenomoics modulator term is nothing but the biologically active compounds that are exposed through forward or reverse chemogenomoics approaches because they bind to and transform specific molecular targets, thus they could be used as ‘targeted therapeutics’[13].
2.1 Forward Chemo Genomics
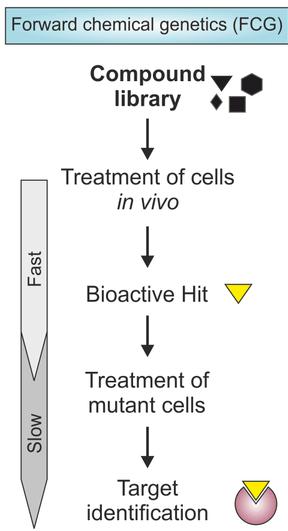
In this approach, compounds are identified on the basis of their conditional phenotypic effect on the whole biological system rather than on the basis of their inhibition of a specific protein target. Lead compound can be classified on the basis on their created phenotypic responses (e.g., cytotoxicity pattern among a panel of cancer cell lines), and such information may smooth the progress of explanation of the compounds mechanism of action which is stated in Figure 1. This type of information is extensively applied in the field of cancer-cell biology[12,14-17]. For the improved understanding the study of cancer chemotherapy and treatment the developmental therapeutics program of the National Cancer Institute (NCI) has started compound screens of cell proliferation using a panel of representative cancer cell lines (NCI60). The anti-proliferative effect of every molecule on every cell line was recorded and the accumulated dataset was analyzed to differentiate various classes of anti-proliferative compounds. This allows researchers to quickly make hypotheses about the mechanism of action of novel cytotoxic agents. Predictive cancer chemogenomics will potentially allow researchers to design more specific clinical trials for achieving better efficacy of novel anti-cancer drug candidates and may ultimately lead to future personalized chemotherapy[18].
|
Figure 1. Flow chart of Forward Chemo Genomics.
2.2 Reverse Chemo Genomics
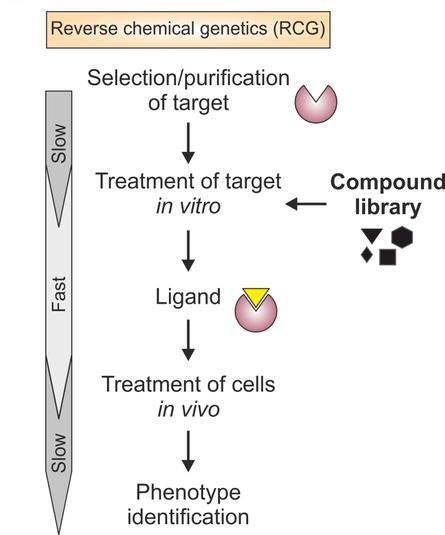
Gene sequences of interest are first cloned and expressed as target proteins in ‘reverse chemogenomoics which is stated in Figure 2. The compound library are screened through high-throughput, ‘target-based’ manner by the called high-throughput screening method with many different bioassays[19-24], which monitor the effects of different compounds on specific targets (such as the ability to bind a protein), on specific cellular pathways[25] or on the phenotype of a whole cell or organism. The assays can be generally divided into cell-free, cell-based and organism assays[26,27]. Several compounds are simultaneously tested for their binding affinity to a wide panel of specific targets are usually called cell-free assay. Universal binding assay is simple, precise, highly automated and compatible with a very high throughput approach in which target-ligand interactions are clearly identified in the absence of confusing variables.
|
Figure 2. Flow chart of Reverse Chemo Genomics.
3 APPLICATIONS OF CHEMO GENOMICS
3.1 Determining Mechanism of Action
Synthetic compounds are less soluble than conventional/traditional (naturally occurring agents) because they have specific structural moiety means the said molecules are mostly bound in various living organism and have extra safety and tolerance factors. chemogenomoics has been used to identify the mechanism of action of drug molecule by studying biological targets. Therefore, this is a resource is used for the development of new molecular entities[28].
3.2 Identifying New Drug Targets
Depending upon the chemogenomoics similarity principle the researchers dock the receptor/enzyme/proteins with ligand library to other members of the same family to identify new targets for the known ligands. The study of drug-receptor complex depends on the availability of an existing ligand library for a same enzyme. Therefore chemogenomoics can be used to identify totally new therapeutic targets.
3.3 Identifying Genes in Biological Pathways
chemogenomoics was used to discover the enzyme responsible for the final step in its synthesis of histidine derivative e.g. Diphthamide is a modified histidine residue found on the translation elongation factor. The first two steps of the biosynthesis pathway leading to dipthine have been known, but the enzyme responsible for the amidation of dipthine to diphthamide remains unknown[29].
3.4 Reference Shallow Methods in Machine Learning for Chemogenomics
A diverse chemogenomoics method has been projected in the last decade. They are differing by the descriptors used to encode proteins and ligands. How similarities are measured between these encoding between proteins and ligands objects, the machine-learning algorithm is used to learn the model and make the predictions. The scientist Jacob and Vert used the Kronecker product of protein and ligand essential part to define the core part associated with the chemogenomic space, i.e., the space of (protein, molecule) pairs. This approach has been successfully applied to DTI prediction. Matrix factorisation approach decompose the (protein, molecule) interaction matrix into the product of two matrices of lower ranks that operate in the two corresponding latent spaces of proteins and molecules[30].
3.5 Computational Chemo Genomics Drug Repositioning Strategy
Drug repositioning is progressively more attractive approach can reduce costs, risks, and time-to-market. As per the literature serve it has been reported to identify novel antimalarial hits by this approach. They have studied the comparative in silico chemogenomics approach to select Plasmodium falciparum and Plasmodium vivax proteins as potential drug targets and analyzed them using a computer-assisted drug repositioning pipeline to identify approved drugs with potential antimalarial activity[31].
4 CONCLUSION
The chemogenomoics approach, where gene sequence information is combined with protein structure and/or models to link to chemical impediments, is designed to completely use the sequence information by considering large families of gene targets at formerly. This largely resembling approach depends on well-established styles similar as combinatorial chemistry, high-outturn webbing, computational chemistry, structural biology and bioinformatics, all of which drive the effective play of information, reagents, styles and know- how as exploration brigades move from one group of targets to the coming. It's still an open question whether a gene family focus is more effective than a ‘traditional’ approach. still, early literature shows that protein kinases and the caspases support opinion that a gene family approach can give a more effective process for generating late-stage development campaigners. Clinical and marketing moxie in specific remedial areas is also of great significance to the success of any new medicine, and traditionally pharmaceutical companies have organized their R&D fret to capture the advantages of this moxie. It's essential that gene family enterprise completely use similar technical knowledge in particular remedial areas while at the same time not limiting the pursuit of targets in other remedial areas. These particular challenges, while complex, aren't invincible with foresight and skillful planning. In this regards we have reported some approaches and operations for chemogenomoics.
Acknowledgements
The authors are grateful to the Management and Principal of K.K.Wagh College of Pharmacy,Nasik for providing necessary facilities.
Conflicts of Interest
The authors declared no conflict of interest, financial or otherwise.
Author Contribution
Amrutkar RD was responsible for studying the conception and design, collecting data, preparing the draft manuscript, and approving the final version of the manuscript. Bhamare VG assisted with data collection and draft editing, while Kapse SN also contributed to data collection.
Abbreviation List
NCI, National Cancer Institute
References
[1] Bredel M, Jacoby E. Chemogenomics: an emerging strategy for rapid target and drug discovery. Nat Rev Genet, 2004; 5: 262-275. DOI: 10.1038/nrg1317
[2] Namchuk MN. Finding the molecules to fuel Chemogenomics. Targets, 2002; 1: 125-129. DOI: 10.1016/S1477-3627(02)02206-7
[3] Caron PR, Mullican MD, Mashal RD et al. Chemogenomic approaches to drug discovery. Curr Opin Chem Biol, 2001; 5: 464-470.
[4] Ambroise Y. Chemogenomic techniques. 2013. Available at: https://archive.md/20130823194737/http://www-dsv.cea.fr/en/institutes/institute-of-biology-and-technology-saclay-ibitec-s/units/molecular-labelling-and-bio-organic-chemistry-scbm/bioorganic-chemistry-laboratory-lcb/transport-and-metabolism-of-iodine-y.-amb
[5] Wuster A, Babu MM. Chemogenomics and biotechnology. Trends Biotechnol, 2008; 26: 252-258. DOI: 10.1016/j.tibtech.2008.01.004
[6] MohdFauzi F, Koutsoukas A, Lowe R et al. Chemogenomics approaches to rationalizing the mode-of-action of traditional Chinese and Ayurvedic medicines. J Chem Inf Model, 2013; 53: 661-673. DOI: 10.1021/ci3005513
[7] Engelberg A. Iconix Pharmaceuticals, Inc.-removing barriers to efficient drug discovery through chemogenomics. Pharmacogenomics, 2004; 5: 741-744. DOI: 10.1517/14622416.5.6.741
[8] Bhattacharjee B, Simon RM, Gangadharaiah C et al. Chemogenomics profiling of drug targets of peptidoglycan biosynthesis pathway in Leptospirainterrogans by virtual screening approaches. J Microbiol Biotechnol, 2013; 23: 779-784. DOI: 10.4014/jmb.1206.06050
[9] Cheung-Ong K, Song KT, Ma Z et al. Comparative chemogenomics to examine the mechanism of action of dna-targeted platinum-acridine anticancer agents. ACS Chem Biol, 2012; 7: 1892-1901. DOI: 10.1021/cb300320d
[10] Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov, 2002; 1: 727-730. DOI: 10.1038/nrd892
[11] Drews J. Genomic sciences and the medicine of tomorrow. Nature Biotechnol, 1996; 14: 1516-1518. DOI: 10.1038/nbt1196-1516
[12] Dafniet B, Cerisier N, Boezio B et al. Development of a chemogenomics library for phenotypic screening. J Cheminform, 2021; 13: 1-12. DOI: 10.1186/s13321-021-00569-1
[13] International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature, 2001; 409: 860-921. DOI: 10.1038/35057062
[14] Drews J, Ryser S. Drug Development: The role of innovation in drug development. Nat Biotechnol, 1997; 15: 1318-1319. DOI: 10.1038/nbt1297-1318
[15] Bhargava H, Sharma A, Suravajhala P. Chemogenomic Approaches for Revealing Drug Target Interactions in Drug Discovery. Curr Genomics, 2021; 22: 328-338. DOI: 10.2174/1389202922666210920125800
[16] Han CK, Ahn SK, Choi NS et al. Design and synthesis of highly potentfumagillin analogues from homology modeling for a humanMetAP-2. Bioorg Med Chem Lett, 2000; 10: 39-43. DOI: 10.1016/S0960-894X(99)00577-6
[17] Ueda H, Nakajima H, Hori Y et al. Action of FR901228, A novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceumno. 968, onHa-ras transformed NIH3T3 cells. Biosci Biotechnol Biochem, 1994; 58: 1579-1583. DOI: 10.1271/bbb.58.1579
[18] Yoshida M, Kijima M, Akita M et al. Potent and specific inhibition of mammalian histone deacetylase both in vivoand in vitroby trichostatin A. J Biol Chem, 1990; 265: 17174-17179. DOI: 10.1016/S0021-9258(17)44885-X
[19] Kwon HJ, Owa T, Hassig CA et al. Depudecin induces morphological reversion of transformed fibroblasts via the inhibition of histone deacetylase. Proc Natl Acad Sci U S A, 1998; 95: 3356-3361. DOI: 10.1073/pnas.95.7.3356
[20] Sandor V, Bakke S, Robey RW et al. Phase I trial of the histone deacetylase inhibitor, depsipeptide (FR901228, NSC 630176), in patients with refractory neoplasms. Clin Cancer Res, 2002; 8: 718-728.
[21] Piekarz RL, Robey R, Sandor V et al. Inhibitor of histone deacetylation, depsipeptide (FR901228), in the treatment of peripheral and cutaneous T-cell lymphoma: a case report. Blood, 2001; 98: 2865-2868. DOI: 10.1182/blood.V98.9.2865
[22] Marshall JL, Rizvi N, Kauh J et al. A phase I trial of depsipeptide (FR901228) in patients with advanced cancer. J Exp Ther Oncol, 2002; 2: 325-332. DOI: 10.1046/j.1359-4117.2002.01039.x
[23] Carducci MA, Gilbert J, Bowling MK et al. A Phase I clinical and pharmacological evaluation of sodium phenylbutyrate on an 120-h infusionschedule. Clin Cancer Res, 2001; 7: 3047-3055.
[24] Gore SD, Weng LJ, Figg WD et al. Impact of prolonged infusions of the putative differentiating agent sodium phenylbutyrate on myelodysplastic syndromes and acute myeloid leukemia. Clin Cancer Res, 2002; 8: 963-970.
[25] Nemunaitis JJ, Orr D, Eager R et al. Phase I study of oral CI-994 in combination with gemcitabine in treatment of patients withadvanced cancer. Cancer J, 2003; 9: 58-66. DOI: 10.1097/00130404-200301000-00010
[26] Kelly WK, Richon VM, O'Connor O et al. Phase I clinical trial of histone deacetylase inhibitor: suberoylanilide hydroxamic acid administered intravenously. Clin Cancer Res, 2003; 9: 3578-3588.
[27] Kubinyi H. Chemogenomics in Drug Discovery. 2006: 1-19. DOI: 10.1007/978-3-540-37635-4_1
[28] Nguyen C, Teo JL, Matsuda A et al. Chemogenomic identification of Ref-1/AP-1 as a therapeutic target for asthma. Proc Natl Acad Sci U S A, 2003; 100: 1169-1173. DOI:10.1073/pnas.0437889100
[29] Playe B, Stoven V. Evaluation of deep and shallow learning methods in chemogenomics for the prediction of drugs specificity. J Cheminform, 2020; 12: 11. DOI:10.1186/s13321-020-0413-0
[30] Rodríguez-Pérez R, Bajorath J. Evolution of Support Vector Machine and Regression Modeling in Chemoinformatics and Drug Discovery. J Comput Aid Mol Des, 2022, 36: 355-362. DOI: 10.1007/s10822-022-00442-9
[31] Ferreira LT, Rodrigues J, Cassiano GC et al. Computational Chemogenomics Drug Repositioning Strategy Enables the Discovery of Epirubicin as a New Repurposed Hit for Plasmodium falciparum and and P. vivax Antimicrob Agents Ch, 2020; 64: e02041-19. DOI: 10.1128/AAC.02041-19
Copyright © 2023 The Author(s). This open-access article is licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, sharing, adaptation, distribution, and reproduction in any medium, provided the original work is properly cited.





 Copyright ©
Copyright ©