Begomovirus: Exploiting the Host Machinery for Their Survival
Vineeta Pandey1, Aarshi Srivastava1#, Niayesh Shahmohammadi2#, Chitra Nehra3#, Rajarshi Kumar Gaur1*, Alireza Golnaraghi4,5*
1Department of Biotechnology, Deen Dayal Upadhyaya Gorakhpur University, Uttar Pradesh, India
2Department of Plant Protection, University of Tehran, Karaj, Iran
3Department of Biosciences, Mody University of Science and Technology, Rajasthan, India
4Department of Plant Protection, Science and Research Branch, Islamic Azad University, Tehran, Iran
5Department of Biodiversity, BoomZista Institute, Vancouver, British Columbia, Canada
#Both authors contributed equally to this manuscript.
*Correspondence to: Rajarshi Kumar Gaur, PhD, Professor, Department of Biotechnology, Deen Dayal Upadhyaya Gorakhpur University, Gorakhpur 27309, Uttar Pradesh, India; Email: gaurrajarshi@hotmail.com;
Alireza Golnaraghi, PhD, Assistant Professor, Department of Plant Protection, Science and Research Branch, Islamic Azad University, Ashrafi-e-esfahani High Way, Hesarak Street, Poonak Square, Tehran 1477893855, Iran; Department of Biodiversity, BoomZista Institute, No. 101, 5325 West Boulevard, Vancouver V6M 3W4, British Columbia, Canada; Email: agolnaraghi@yahoo.com
Abstract
Begomoviruses are the most prevalent plant-infecting virus genus, identified by a circular single-stranded DNA genome. The proteins encoded by these viruses are multifunctional and effectively modulate the host's cellular response during infection. Begomovirus-host interactions alter host gene expression patterns, redirect signaling pathways, disturb protein degradation pathways, and reprogram cellular metabolism to establish a successful infection. This review summarises known studies about the genome organisation of begomoviruses and altered cellular processes during infection at a molecular level. In addition, we emphasise proteins involved in intercellular trafficking of the viral genome for systemic infection and counteracting host small interfering RNA silencing and discuss different approaches for virus management based on the virus’s offensive and the host’s defensive strategies.
Keywords: begomovirus, virus-host interaction, host responses, virus infection strategies, management
1 INTRODUCTION
Geminiviridae viruses are small, non-enveloped microorganisms containing circular, mono- or bipartite, single-stranded DNA (ssDNA) genomes (2.5-5.2kb). Based on host range (monocots or dicots), insect vectors (leafhoppers, treehoppers, whiteflies, and aphids), genome organisation (mono- or bipartite), and phylogenetic relationships, the Geminiviridae family is classified into fourteen genera: Begomovirus Becurtovirus, Curtovirus, Citlodavirus, Capulavirus, Eragrovirus, Grablovirus, Mastrevirus, Maldovirus, Mulcrilevirus, Opunvirus, Turncurtovirus, Topilevirus, and Topocuvirus. Out of all these genera, Begomovirus is the most widespread genus, with >440 species, and has been emerging as a serious threat to crop production across the world for the past few decades. Begomoviruses are transmitted by whitefly vectors and are present in both the old world (mainly mono- and rarely bipartite genome viruses) and the new world (mainly bipartite with some monopartite genome viruses)[1]. The whitefly Bemisia tabaci (Genn.) is a complex cryptic species, and the virus-vector binding depends on specific amino acids present in the viral coat protein (CP). Begomovirus-encoded proteins are multifunctional and effectively regulate host cellular processes to establish a successful infection. Plant-virus interactions might change various host phenomena like the expression of genes, signaling patterns, cellular metabolic pathways, defence mechanisms, and suppression of RNA signaling pathways, resulting in increased host susceptibility to viral infection[2]. Molecular interactions during infection involve an array of host proteins and 5-7 begomovirus-encoded proteins. Viral proteins are complex in nature and interact dynamically with various plant-related proteins to overcome plant defence strategies[2].
Although begomoviruses infect only dicot plants but a recent study explores its infectivity in monocot plants too[3]. These viruses have caused significant yield and quality losses in crops, affecting both food and nutritional security globally[4]. This article reviews the current knowledge regarding molecular mechanisms involved in geminivirus-host interactions, virus intracellular trafficking, and alternation and modification of host genes against virus infection. The mechanism and biological roles of virus-encoded proteins in manipulating host-mediated responses and the vector transmission process are also summarised in the review to understand the offensive strategies for virus management.
2 GENOME ORGANIZATION AND REPLICATION
2.1 DNA-A and DNA-B
Monopartite begomoviruses only consist of DNA-A (~2.5kb), while bipartite viruses have two circular genomic components, DNA-A and DNA-B (each ~2.5kb in size)[5]. Both DNA elements share a common region that serves as the replication origin and aids in bidirectional transcription. Begomovirus DNA-A can multiply autonomously to create virions, but DNA-B aids in systemic infection. The open reading frames (ORFs) present in DNA-A of monopartite genomes are almost like the DNA-A component of bipartite viral genomes[1]. The virion-sense strand of DNA-A encodes both the CP (ORF AV1/V1), which encapsidates ssDNA and might be functional in the movement of the virus, and the AV2/V2 protein (ORF AV2/V2) that is also associated with virus movement. This AV2/V2 ORF is absent in the new world begomoviruses. The anti-sense strand of DNA-A ORF AC1/C1 encodes, replication-associated protein (Rep), ORF AC2/C2 encodes, transcriptional activator protein, ORF AC3/C3 encodes replication enhancer protein, and ORF AC4/C4 encodes AC4/C4 protein[6]. On the virion-sense strand, DNA-B encodes a nuclear shuttle protein (NSP), ORF BV1 and on the complementary-sense strand, movement protein (MP), ORF BC1. Both NSP and MP are essential to improve viral DNA inter- and intracellular transport as well as systemic infection[7]. Furthermore, these two proteins allow nucleocytoplasmic translocation via nuclear pores and cell-to-cell migration of viral DNA via plasmodesmata, respectively (Figure 1).
|
Figure 1. Genomic organization of monopartite and bipartite Begomovirus species. Arrows show ORFs in virion-sense and anti-sense strands of DNA-A and DNA-B segments.
2.2 Satellites
Three different types of circular ssDNA satellites have been reported to be associated with begomoviruses, where the DNA-A element works as a helper DNA and gives machinery to the satellite components for replication and transcription of respective ORFs. These satellites are named alphasatellites (α), betasatellites (β), and deltasatellites (δ)[8] (Figure 2). Geminivirus alphasatellites are ~1.4kb in size, the adenine-rich circular DNA molecules that encode a Rep protein. Alphasatellites do not rely on helper viruses during replication, but they do for encapsidation[9]. Alphasatellites are believed to originate from nanovirus Rep-encoding elements and can replicate autonomously in host plant cells, however, the precise contribution of alphasatellites to viral pathogenicity is unknown. Although alphasatellites have been associated with reduced infection symptoms and decrease in viral DNA and betasatellite titers[10], they also play important roles in the suppression of transcriptional gene silencing (TGS) and post-transcriptional gene silencing (PTGS). Recently, it has been shown that some alphasatellites of nanoviruses affect neither the symptom severity nor the overall helper virus accumulation but rather change the relative amounts of the genomic segments and the rate of plant-to-plant transmission[11].
|
Figure 2. Schematics depicting general genome organization and features observed in betasatellites, alphasatellites and deltasatellites.
Moreover, betasatellites are circular ssDNA molecules (~1.35kb) often found in association with monopartite begomoviruses and infect a wide range of host plants[12]. For encapsidation, replication, and movement, betasatellites require helper viruses for proper functions, but play essential roles in symptom development and maintaining disease in the field. Betasatellites show no sequence homology with cognate helper viruses other than a conserved nonanucleotide (TAATATTAC) containing a stem-loop structure. This conserved sequence is important for the trans-replication of betasatellites by their helper DNA-A. Betasatellites encode a multifunctional βC1 protein (~118aa) that participates in diverse cellular events like virus movement and suppression of gene silencing, and may also be involved in enhancing disease symptoms, virus titer, and whitefly transmission[13]. Subsequently, deltasatellites are ~0.7kb in size and do not encode any protein. They have been identified in the presence of old world monopartite begomoviruses, new world bipartite begomoviruses, and sweepoviruses[8]. Deltasatellites rely on their begomovirus companion for reproduction, mobility, and transmission. In most cases, these satellite molecules do not affect the disease symptoms, but in some cases, they have been found to show effects on increasing or reducing viral DNA accumulation[14,15].
3 VECTOR TRANSMISSION AND VIRUS-VECTOR INTERACTIONS
Begomoviruses are transmitted in host plants via a cyclic triad formation and interaction among the host, vector (whitefly), and virus. Virus particles acquired by whiteflies from infected leaves during their feeding process, move to the alimentary canal and then to the midgut and hemolymph of whiteflies. Ultimately, viruses are collected in the salivary gland of the vector for future transmission via the feeding process[16]. In the midgut of the whitefly, virus CP interacts with GroEL and heat shock protein 16, which include cyclophilins[17], easing virion movement and translocation motion across membranes and aiding the triad interaction. In addition to that, all these interactions among proteins happen in the midgut of whiteflies and promote viral movement from the midgut to haemolymph via endocytosis, which enables the ejection of viruses into host plants with salivary secretion[18].
Biological and molecular traits can be used to differentiate vector biotypes. The B biotype is the primary driver of begomovirus emergence, but new evidence suggests that the situation is more complicated, with biotypes differing in transmission efficiency, competing and displacing one another, and interacting mutually with begomoviruses. This could potentially be a result of global climate change, bringing vectors and viruses from temperate and tropical regions together. Thus, this can stimulate the formation of new combinations of geminiviruses and satellites through mixed infections. For instance, a more complex situation being studied in China involves a cooperative interaction between begomoviruses and B. tabaci biotypes, which first appeared in the mid-1990s and replaced the indigenous biotype (ZHJ2). This results in the emergence of the tomato yellow leaf curl China virus (TYLCCNV) and a betasatellite. Moreover, the tomato yellow leaf curl virus (TYLCV) was introduced in 2000 and had become prevalent by 2012. However, the expansion and emergence were found to be particularly mediated by the Q biotype vector because of its greater survival and fecundity than the B biotype[19]. Thus, due to direct vector manipulation by the virus, altered feeding behaviour, and indirect vector affinity for TYLCV-infected plants, this mutualistic interaction favoured both the vector and the virus. It will be of interest to determine the factors in begomovirus-infected plants that are detrimental to one biotype but favourable to another biotype and to identify whether mutualism prevails in other B. tabaci begomovirus associations or not. The impact of such mutualism would help investigate begomovirus prevalence and diversity[20].
4 CELLULAR REPROGRAMMING
4.1 Alteration of Gene Expression in Host
Begomovirus encodes a Rep protein that only initiates replication, so the virus must rely on host replication machinery to replicate its genome. The virus accumulates DNA replication machinery by rerouting host gene expression to compensate for the lack of host DNA polymerase and associated factors in differentiated host cells. Begomovirus proteins interact with host DNA replication and transcription machinery, cell division, and metabolic pathway proteins, including defence and stress-related proteins (Table 1). Rep interferes with host DNA replication by binding with host recombination/repair proteins, DNA polymerase complex[21], and ssDNA binding proteins[8]. Viral proteins interact with host transcription factors to regulate gene expression in host cells. Various transcription factors like WRKY, NAC, MYB, and leucine zipper have been found to control viral infection in host-virus relations[22,23]. Cabbage leaf curl virus (CaLCuV) infection of Arabidopsis regulates cell cycle genes and directs passage further into the endocycle (Figure 3A). Begomovirus proteins interact with plant protein kinase to modify signaling pathways. More activity of receptor-like kinases is needed in addition to the infection process[24]. A pathogenicity determinant protein (C4) of tomato leaf curl Australia virus interacts with the Shaggy-like protein kinase of the brassinosteroid signaling pathway[25], and an AC2 encoded protein deactivates the adenosine kinase (ADK) and sucrose-nonfermenting 1 related protein kinase to restrict the host's basal immune response[26]. Further, it has been found that several genes in host plants are expressed during regulating systemic acquired resistance, while the other genes are engaged in cytoplasm-to-nucleus and cell-to-cell virus trafficking. Investigations on geminivirus-host interactions indicate that these viruses can modulate the host metabolism by triggering the transcription of host genes in mature tissues to accumulate host replication factors[27]. Therefore, this demonstrates that alteration in plant development activities during infection by viruses in host cells delivers a molecular relationship between metabolism and inherent vulnerability to viral infections.
Table 1. List of Key Plant Proteins Showing Altered Expression as Resistance Response Against Viral Infection
Sr. No. |
Proteins |
Function |
Effects on Host Systems Upon Viral Infection |
Ref. |
1. |
Receptor-like kinase (RLKs) |
Instigate an array of defence related signal cascades or PTI (PAMP triggered immunity) and thereby impart resistance |
Up-regulation of RLKs triggers the downstream defence responses involving complex signaling cascades |
[24] |
2. |
Mitogen-activated protein kinases (MAPK), serine threonine-protein kinase, lectin protein kinase, tyrosine-protein kinase |
Signaling molecules |
Act as signaling molecule for the upstream activation of immune responses against YMV |
[28] |
3. |
Jasmonate-ZIM-domain (JAZ) protein resembling domain in the PPD2 (plant-specific putative DNA-binding proteins) |
Binds to the DNA elements in the viral CP promoter region |
Regulates the virus multiplication |
[29] |
4. |
3-epi-6-deoxocathasterone 23-monooxygenase |
A brassinosteroid (BR) |
Altered expression |
[30] |
5. |
Respiratory burst oxidase homolog protein |
Signaling protein |
Altered expression |
[31] |
6. |
WRKY, C2-H2 Zinc finger, Leucine zipper |
Transcription factors |
Altered expression to regulate virus infection |
[32,33] |
7. |
NAC |
Transcription factor |
Negatively affect viral DNA replication |
[33] |
8. |
Pathogenesis-related (PR) proteins, defensin-like proteins |
Systemic acquired resistance related proteins |
Up regulation induces defence related genes in host |
[34] |
9. |
GTP-binding proteins |
Receptor proteins |
Induce defence related genes in host upon infection |
[35] |
10. |
Resistance (R) proteins (glutathione S-transferase, heat shock protein (HSPs), and ferredoxin) |
Activate hypersensitive response (HR) as resistance mechanism |
Downstream synthesis of a range of R-proteins leading to the resistance response |
[36,31] |
11. |
Secondary metabolites |
Develop plant immunity |
Activated production upon infection |
[37] |
12. |
Photosynthesis related proteins |
Photosynthetic activity |
Except chlorophyllase-2 all other chlorophyll- and carotenoid-related proteins are downregulated and cause yellow mosaic symptoms |
[38] |
13. |
Phenylalanine ammonia-lyase (PAL) |
Contribute to several stress response related pathways |
Up-regulated in response to CCYV (cucurbit chlorotic yellows virus) infection |
[33] |
14. |
RNA-dependent RNA polymerase (RDR6), Argonaute (AGO) family proteins |
RNA silencing pathways proteins |
Activated synthesis upon infection |
[39] |
15. |
Endochitinases |
Correlated with host resistance |
Up-regulation |
[31] |
16. |
Ubiquitin-proteases |
Protein degradation pathway enzymes |
Up-regulation |
[31,40] |
17. |
Iron-Sulfur Cluster (ISC) assembly protein |
Required for the maturation of RNase L Inhibitor 1 (RLI1) for maturation of virus particles |
Down- regulation |
[41] |
18. |
Asymmetric Leaf 1 (AS1) |
|
Decreases expression of many JA-responsive genes in response to TYLCCNV |
[42] |
19. |
SNF1 (sucrose-nonfermenting 1)-related kinase |
Key role in the regulation of cellular stress responses and carbon metabolism |
Kinase activity, including autophosphorylation, will be abolished |
[43] |
20.
|
Brassinosteroid insensitive 1- associated kinase 1 (BAK1) and a PERK-like kinase |
Receptor-like kinases |
Suppression of PAMP recognition receptors and PTIs |
[44] |
21. |
AS2, AtNSI, H3, NIG and G3BP, AtWWP1 |
Pro-viral and antiviral proteins |
Suppression of post- transcriptional gene silencing (PTGS) |
[45] |
22. |
Calmodulin-like proteins |
Plant endogenous gene-silencing suppressors |
Regulate downstream proteins upon infection |
[46] |
23. |
SIUBC3 |
Ubiquitin-conjugating enzyme |
Disrupting the proteasomal degradation pathway and altering plant hormonal signaling cascades |
[47] |
|
Figure 3. Begomovirus-plant interactions. A: Changes in host genes expression; B: miRNA-phytohormone interplay; C: Changes in cellular metabolism (protein degradation).
4.2 Modifications in Signaling Pathways
During infection, begomoviruses penetrate and establish themselves successfully in the cell, and this infection will then spread to neighbouring cells, causing systemic infection throughout the plant, and also affect the host stress-signaling pathways. Phytohormones control various physiological responses in plants, which will be changed during viral infection[40]. Stress responses activate several hormone-regulated immunological signal transductions[48]. Jasmonic acid (JA) plays an important role in defence responses by limiting photosynthesis, and plants utilise cell division to keep a balance between growth and defence[49]. One such example is in Vigna radiata, where lipoxygenase (LOX) and Jasmonate-ZIM-domain (JAZ) proteins have been detected as up-regulated in mungbean yellow mosaic virus (MYMV) resistant plants in comparison to susceptible plants. A JAZ protein that matches the domain seen in crop-specific putative DNA-binding proteins was found to bind with DNA components found in the viral CP gene promoter that controls virus replication[36]. Colnelic and colnelenic acids, which are products of the LOX pathway, trigger inhibitory activity after accumulation in tobacco mosaic virus-infected potato leaves. The miR319/TCP regulates the JA biosynthetic gene LOX2 and modulates the miR164/CUC node as a component of the auxin-signaling network (Figure 3B)[50].
In the case of viral infection, these cascade responses may be regulated at the transcriptional level in the plant. Though the relationship between modified gene expression and disease symptom development is not fully understood, it is supposed that various viral proteins, which are called pathogenicity determinants, are involved in symptom development[32]. Virus infection can affect the synthesis of many hormones like auxins, ethylene, cytokinin, and gibberellins by differentially regulating the expression of associated genes[32,38].
4.3 Modifications in Host Protein Degradation Pathways
During infection, begomovirus proteins can obstruct plant pathways for protein breakdown, thereby regulating protein metabolism in host cells. Numerous reports explain the molecular defence mechanisms used by hosts, such as sumoylation, ubiquitination, and protease-mediated degradation to modify plant growth during plant-virus interaction (Figure 3C)[9,51]. Ubiquitination plays a role in plant defence, including virus resistance. Tomato UBA1 has been shown to interact with tomato yellow leaf curl Sardinia virus (TYLCSV)-derived C2, impairing the derubylation action of the COP9 signalosome (CSN) complex and therefore constraining the action of the Skp1/Cullin1/F-box (SCF) complex, which is associated with the ubiquitylation pathway of the host plant[52]. This TYLCSV C2-induced inhibition of the SCF complex was also considered to hurt the jasmonate signaling pathway. A similar example is beet severe curly top virus (BSCTV)-derived C2 interfering with host-derived E3 ligase which leads to inhibition of S-adenosylmethionine decarboxylase proenzyme 1 degradation and subsequent suppression of viral gene silencing via methylation-dependent mechanisms[53]. BSCTV-C4 activates E3 ligase, which deteriorates cell cycle regulators and obstructs cell cycle-related pathways. TYLCSV, African cassava mosaic virus (ACMV), and tomato golden mosaic virus (TGMV) encoded Rep proteins bind with the E1SUMO-conjugating enzyme and impair sumoylation in plant cells. TYLCSV infection can directly alter genes involved in post-translational modifications such as ubiquitination, rubylation, acetylation, and phosphorylation, which are possibly due to modifications in host machinery in response to infection. The Nicotiana benthamiana system has been applied to monitor TYLCSV infection and subsequent altering or silencing of host genes. To accomplish this, researchers the 2IRGFP N. benthamiana transgenic line, which allows them to examine the virus-induction of host genes and their silencing. The TYLCSV Rep protein precisely identifies the cassette that is flanked by intergenic region during infection and starts replication, and green fluorescent protein transgene expression is highly effective. The system serves as an important tool for tracking infection growth in plant tissues in both space-time in a simple, visual, reliable, and non-invasive manner[54].
4.4 Modifications in Cellular Metabolism
Begomoviruses multiply in host cells by altering host cellular metabolic processes like nutrient rerouting and changing cell wall structure and synthesis[55]. Tomato plant transcriptomics analysis following tomato leaf curl New Delhi virus infection revealed that virus infection caused changes in 77 known pathways generally related to decreasing photosynthesis rate, increasing respiration rate, and affecting sugars, starch, carbohydrates and amino acids accumulation and metabolism. Thus, this altered physiology possibly arise as symptoms. Similarly, during viral infection, the altered levels of acetyl Coenzyme and phosphoenol pyruvate phosphate transcripts were observed, indicating modifications in glycolysis and the tricarboxylic acid (TCA) cycle[32,54]. Tomato TYLCSV infection can affect only a small cell count inside vascular tissues and cause transcriptional changes. This was demonstrated experimentally upon mature leaves' midribs, which were naturally abundant in vascular tissues, and it was found that a total of 1,398 genes were significantly down-regulated, while 2,206 were significantly up-regulated. Among those genes that were involved in the silencing of chromatin by transcription and methylation, primary and secondary metabolism, along with phosphorylation, were down-regulated, while those that were up-regulated included nucleic acid metabolism, the ubiquitin-proteasome pathway, hormone responses and metabolism, and autophagy[56]. Further, C2 of beet curly top virus (BCTV) and AC2 of TGMV have been shown to make contact SNF1- related kinase (SnRK1), a serine-threonine kinase that is important for cellular stress responses and innate defence mechanisms that get inactivated due to geminivirus infection. It has been shown that AL2 and L2 interrelate with SNF1-related kinase, and consequently, kinase activity, including autophosphorylation, will be abolished[43]. The findings also show that transgenic N. benthamiana and N. tabacum var. Samsun plants expressing truncated and full-length AL2 and L2 genes, respectively, display a boosted vulnerability phenotype during inoculation with RNA or DNA viruses due to the inhibition of metabolic responses mediated by SNF1[43]. The other modification was found to elevate cellular cytokinin levels due to ADK inhibition by C2 of spinach curly top virus (SCTV) and AC2 of TGMV, which resulted in an enhanced susceptible phenotype[57]. Another study talks about a highly conserved protein complex, i.e., the CSN, which is comprised of eight subunits (CSN1-CSN8), where CSN5 is the only catalytic subunit that gets into interaction with the C2 gene of TYLCSV during infection and destabilises the cellular ubiquitination machinery[58]. Moreover, the CSN5 subunit is also associated with the deregulation activity, which gets inhibited when there is an association between CSN5 and C2 of TYLCSV. Similarly, C2 of TYLCV was found to down-regulate terpene synthesis, which plays a significant role in plant defence against abiotic and biotic stresses[59]. In the case of papaya leaf curl China virus, it interacts with the ubiquitin moiety of RPS27A, a fusion protein that suppresses proteasome-mediated ubiquitination and decreases JAZ1 degradation, thus interfering with Jasmonate signaling[60].
5 INTRACELLULAR TRAFFICKING
To establish a successful systemic infection in plants, viruses must transfer their genomes among both the cells and the plant. So, host factor detection and viral proteins involved in the motion of the begomovirus ssDNA genome might help in uncovering the mechanism behind viral genome movement and establishing antiviral strategies. The CP gene of monopartite viruses, and NSP of bipartite ones, use host nuclear export machinery to enhance viral genome migration between the nucleus and the cytoplasmin in plant cells. NSP assists with viral MP to translocate viral DNA transfer from one cell to another. CP and NSP bind with both ssDNA and double stranded DNA (dsDNA) in a sequence-independent manner. NSPs of bipartite begomoviruses also connect with NSP-interacting GTPase (NIG) in the cell under transient expression and transmit the viral protein between the nucleus and the cytoplasm by modifying NSP nucleocytoplasmic shuttling. Hence, it was found that NIG acted as a constructive contributor to geminivirus infection and served as a cofactor in the cortical cytoplasm to facilitate MP-NSP interaction[61]. It was shown that overexpression of NIG increased virus susceptibility in transgenic plants, which means it is a positive contributor to the infection process. Begomoviruses replicate their ssDNA in the nucleus of infected cells via dsDNA intermediates. This newly synthesised ssDNA has three options: it can just re-enter the replication cycle or bind with CP (in monopartite begomoviruses) or NSP (in bipartite begomoviruses). The interaction of NSP and viral DNA facilitates intracellular mobility from the cell cytoplasm, mostly through exportin-like receptors. NIG encounters NSP first at the nuclear pore complex's cytosolic face and reroutes the NSP-viral DNA complex to the cell's periphery, where NSP is changed by MP. As a result, NIG may enable directionality in the intracellular mobility of viral DNA, and association and dissociation of NIG-based complexes might be controlled by NIG GTPase activity. MP mediates the transportation of the complex NSP-viral DNA and/or viral DNA to the next cell through plasmodesmata.
In monopartite begomoviruses, CP mediates the translocation of dsDNA that moves from cell to cell and over long distances in the plant. CP was also reported to bind viral genomic DNA by the N-terminal zinc finger motif[62]. A model was proposed for CP-mediated viral DNA migration from cell to cell. According to that model, CP enters the nucleus via nuclear localization signals and binds with ssDNA to export it from the nucleus to the cytoplasm. Another viral protein, V2, which is present at the nuclear periphery, also aids in the nucleus's export of the CP-DNA complex. This CP-DNA complex, along with V2, may be transported to the cell periphery with the involvement of other viral proteins and host factors, followed by transport via plasmodesmata. Although some progress has been made in the recognition of proteins involved in viral genomic DNA translocation, the transport mechanism and the precise potential mechanisms are still unknown. Furthermore, the MP of begomoviruses contributes to expanding the pore size of plasmodesmata and dynamically transmitting the viral nucleic acid into the adjoining cell. Since viruses are too large to fit through an unmodified plasmodesma and to be easily diffused through the cytoplasm of the cell, the finding suggests that it is not only MP but, in addition to this, other viral proteins also participate in the successful intracellular movement of viruses. Accordingly, the NSP of begomoviruses assists in the intracellular transmission of viral DNA between the nucleus and cytoplasm and collaborates with the MP for the cell-to-cell translocation of viral DNA to healthy cells. NSP, on the other hand, acts as a determinant of virulence that overcomes antiviral protection toward begomoviruses. Similarly, this protein also hijacks compatible host functions and encourages the intracytoplasmic and nucleocytoplasmic transfer of viral DNA[63].
6 SUPPRESSING THE HOST DEFENCE SYSTEM
In plants, the RNA silencing mechanism is primarily immune responsive upon pathogen infection. Begomoviruses infection initiates RNA silencing processes in host cells. RNA silencing effector molecules are mainly small interfering RNAs (siRNAs) and microRNAs (miRNAs), which regulate gene expression of both plant and viral genes in a sequence-independent manner[64]. In defence against the host immune response, viruses have evolved viral suppressor proteins (VSRs). Initially, the AC2 proteins of ACMV and TYLCV were shown to reverse host-silencing pathways. Further, it was reported that the AC2 protein had DNA binding domains and active nuclear localization signals (NLS), which are necessary for suppression activity; this protein also involves in the transactivation of viral promoters. Interestingly, begomovirus AC2 proteins do not bind with small RNAs, which is significantly different from RNA viruses VSRs, as they do bind with sRNAs. TGMV AL2 protein was shown to bind with SnRK1, serine/threonine kinase (SNF), proteins that also participate ADK[65], which are key regulators in methylation and metabolic processes in host cells.
According to research, multiple viral infections among the viruses lead to synergism that causes more severe disease symptoms in plants. The VSR protein, derived from viral ORF AC4, was discovered to play an important role in this synergism process. Similarly, when AC4 interacted with ACMV Cameroon strain ACMV-(CM) and East African cassava mosaic virus (EACMCV) AC2, symptom severity increased, indicating a complementation effect of these two proteins. Moreover, AC4 influences the RNA's downstream silencing pathway because it binds single-stranded small RNAs but not double-stranded (dsRNAs). AC5 protein is encoded by a bipartite begomovirus and acts as a VSR by interfering with dsRNA biogenesis[66]. The V2 protein has also been demonstrated to decrease RNA silencing in single-partite viruses TYLCV and TYLCCV by acting on the multiplication of the strength of the silencing signals and decreasing TGS[67].
Consequently, the cotton leaf curl Multan virus (CLCuMV) and its associated cotton leaf curl Multan Β-satellite (CLCuMB) were studied for their ability to cause cotton leaf curl disease (CLCuD); it was discovered that CLCuMV alone was incapable of effectively blocking silencing. In the absence of the betasatellite, however, gene silencing was severely suppressed. Similarly, CLCuMV and cotton leaf curl multan alphasatellite (CLCuMA) were studied in an N. benthamiana system to test the ability to conquer gene silencing. The results showed that silencing was not disrupted in any way when infections involved CLCuMA, even though the alphasatellite was shown for the first time to be a target of RNA silencing, stimulators to produce specific small siRNAs, and effectors of silencing in plants. Subsequently, the capability and capacity of all CLCuMV and CLCuMB-encoded proteins that restrict RNA interference (RNAi) and the relative performance of their suppression activities were evaluated and compared. The results showed that the V2, C2, C4, and βC1 proteins had suppressor activities, with the V2 showing a robust activity. In addition, V2, C4, and βC1 were also shown to have the capability to bind RNA and distinctive specificities[68]. However, the exact mechanism of suppression of RNA silencing by betasatellites is poorly understood, but betasatellite-encoded βC1 has been shown to interact with ssDNA/dsDNA autonomously and remain present in the nucleus. Some reports are showing that βC1 encoded by various begomoviruses betasatellites are acting as VSR[11]. So, begomoviruses have developed various strategies, including viral suppressor proteins, to counter-defend against the host RNAi response by effectively causing gene silencing through virus encoded gene silencing proteins, which allows them to effectively invade plants[69]. These multiple suppressors act at different steps in silencing pathways.
7 INTERACTING WITH THE HOST IMMUNE SYSTEM
Most plants are resistant to most plant pathogens, which can be the result of host-non-host interactions or due to host defence systems. Plants have evolved a range of rapid and efficient defence responses against a wide variety of pathogens, including fungi, bacteria, nematodes, and viruses. Common aspects of host conditioning carried out by most DNA and RNA viruses involve molecular interaction with different host proteins for suppression of their defences. In general, plant defence can be divided into three levels: effector-triggered immunity (ETI), pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), and RNA silencing[70]. The ability of geminiviruses to confront immune systems is usually correlated with their broad host spectrum[71].
Plants use an innate immune pathway to recognise viral effectors and limit viral infection. This innate immune system includes two levels of detection systems to activate defence[72]. In the first level of defence, PTI detect and recognise PAMPs by surface-localized pattern recognition receptors (PRRs)[73]. ETI is a secondary defence pathway that involves resistance proteins (R) with nucleotide-binding leucine-rich repeat (NB-LRR) domains that recognise-directly or indirectly-viral effectors. To identify invading viruses and initiate appropriate defence responses, hosts have evolved signaling pathways. In contrast, viruses have developed counter-defence strategies that can suppress the host defence system and appropriate host cellular resources, leading to disease[42].
signaling pathways, including salicylic acid (SA) and JA, regulate developmental processes and plant responses and are both DNA and RNA virus-triggered. During begomovirus infection, genes implicated in the SA and JA pathways are generally silenced[24]. The βC1 gene from TYLCCNV inhibits the expression of many JA-responsive genes by interacting with asymmetric leaf 1 (AS1)[42], whereas AC2 from ACMV activates JA-responsive genes in transgenic tobacco plants[74]. The brassinosteroid (BR) signaling pathway influences growth and responses to biotic and abiotic stress[75]. Furthermore, the C4 gene encounters brassinosteroid-insensitive 2, bringing negative regulation of BR signaling, which changes the plant's development by interfering with multiple hormonal pathways[76].
The bipartite begomovirus NSP, which involves the movement of viral DNA from the nuclear pore complex and connects the nucleus to the cytoplasm, has been shown to interact with host proteins, allowing the virus to suppress or avoid host defence responses. NIK1 has been shown to play a remarkable role in the defence response against geminiviruses[77]. The NIK1 kinase domain is phosphorylated at the threonine-474 residue when it oligomerizes, which facilitates phosphorylation of the RPL10 to bind to gene L10-interacting MYB domain-containing protein[77,78] and suppresses the expression of genes in translational machinery[79]. Therefore, during virus infection, the viral mRNAs are simply unable to bypass the plant cells' translational control mechanisms; as a result, they are not efficiently translated and cause infection[80]. Overexpression of NIK1 has also been shown to decrease viral infection and its symptom development in tomatoes. Begomoviruses overcome this defence strategy through NSP-NIK1 interactions that hinder the phosphorylation of the threonine-474 residue to inhibit the activation of NIK1. This subsequently prevents the transduction of antiviral signals and enhances susceptibility to begomovirus infection[81]. The NSP-NIK association is shared by begomovirus NSPs and NIK homologues from various hosts. The study suggests that the NIK1 immune receptor-like kinase could be used to generate broad begomovirus resistance[80].
NSP can form an NSP-associated kinase containing two receptor-like kinases, the Brassinosteroid Insensitive 1 (BAK1) and a PERK-like kinase[82]. This complex is thought to have a host defence-reduction capability by suppressing BAK1 as a co-receptor of many PRRs, which detects certain PAMPs and triggers or enhances PTI[44]. In addition to kinase receptors, NSP interacts with other kinds of plant proteins, like pro-viral proteins asymmetric leaves 2, AtNSI, H3, and NIG[83], and antiviral proteins such as G3BP and AtWWP1[45], leading to suppression of PTGS and increasing susceptibility to begomovirus[63,84]. Interaction of the AC1 gene of TYLCCNV with AS1 causes changes in leaf development and the appearance of disease symptoms[39]. Begomovirus infection has been shown to induce a hypersensitive response (HR) as well. C4 interacts with the hypersensitive induced reaction 1 (HIR1) gene to inhibit its function and mediate HR by stimulating its degradation and impairing HIR1 self-oligomerization[85].
Betasatellites can reduce TGS and PTGS activity[9]. linked with TYLCCNV binds with endogenous gene-silencing repressors such as calmodulin-like proteins, in N. benthamiana, which when activated, alters the host signaling mechanism and enhance the viral disease[46]. In vivo studies also showed an interaction of the myristoylation-like motif present in βC1 of the cotton leaf curl Gezira betasatellite with SIUBC3 as an ubiquitin-conjugating enzyme. This interaction leads to the repression of the plant ubiquitination system, changing plant hormone signaling cascades, and affecting the proteasomal destruction pathway[47]. Interruption of different subunits of the plant ubiquitination system by CLCuMuB-βC1 results in an increase in the amount of cotton leaf curl Gezira virus (CLCuGV) DNA accumulation and severe disease symptoms of the virus infection because of the repression of the JA and gibberellic acid pathways[86].
8 STABILITY AND PATHOGENICITY DETERMINANTS
For the successful viral infection of the plant host, pathogenicity indicators are viral proteins that have evolved to break host resistance through a network of protein interactions[87]. Many studies have confirmed that AC2, AC4, AV2 (pre-CP), and NSP are major pathogenicity determinants that also act as viral RNAi suppressors. These proteins require post-translational modifications for their activity in infection and counter-defence. Therefore, they use the cellular systems, including the hijacking of the host PTM machinery, as observed in the tomato leaf curl Yunnan virus-C4 and CaLCuV-NSP proteins, both of which necessitate phosphorylation to function[1].
Among geminivirus proteins, AC4/C4 is the lowest conserved, and its activity differs among geminiviruses. This protein is a significant Curtovirus pathogenic determinant as well as some monopartite begomoviruses, involves in symptom development. Moreover, inactivation of C4 diminishes viral infection and symptom development[30,88]. Even though AC4 of the bipartite begomovirus TGMV has no apparent effect on symptom growth, the AC4 protein of another bipartite begomovirus, tomato leaf curl Puangdong virus (ToLCPalV), is a viral pathogenicity determinant, a disease establishment factor, and an RNA silencing suppressor. Transient expression of the ToLCPalV AC4 in N. benthamiana by the potato virus X (PVX) vector resulted in the enhancement of ToLCPalV symptoms like downward leaf puckering, leaf curling, and tissue necrosis[89]. This result was also true for tomato leaf curl Guangdong virus (ToLCGdV) and MYMV because their C4 suppressed TGS and PTGS by interacting with the barely any meristem 1 gene in the transgenic N. benthamiana line 16c[90,91]. For ACMV and Sri Lankan cassava mosaic virus (SLCMV), This protein also inhibited TGS by binding exclusively to miRNAs and siRNAs, preventing miRNA-mediated degradation of target mRNAs[90]. This action, however, was not observed in EACMCV or Indian cassava mosaic virus (ICMV)[87]. AC4 association with arabidopsis shaggy-like kinase family members modulates the BR signaling pathway, which results in plant abnormal development[92,93].
Other begomoviral proteins like BCTV V2, as well as ToLCNDV and bean dwarf mosaic virus (BDMV)-NSP, can induce systemic symptoms and necrosis associated with HR when expressed from the PVX vector in each of N. benthamiana, N. tabacum, Solanum lycopersicum, or Phaseolus vulgaris plants. These proteins are both virulence determinants and target host defence responses, so their pathogenicity is independent of their ability to suppress PTGS. Researchers have recently found some additional phylogenetically conserved ORFs, including V3 in geminiviruses, particularly in begomoviruses with unique subcellular localizations and characteristics. This gene is required for infection in N. benthamiana and tomato, and it encodes a Golgi-localized protein that functions as a PTGS and TGS suppressor[94].
For many monopartite viruses, DNA-A alone is sufficient for infection and disease symptom generation[46], but the presence of betasatellites induces serious symptoms in grown crop species, apart from some non-cultivated species, and increases the virus titer. The interaction of TYLCCNV/TYLCCNB with four genes NbHEN1, NbDCL3, NbDRM2 and NbAGO4, when studied on the N. benthamiana system, was discovered to be engaged in RNA-directed DNA methylation (RdM), increased viral DNA accumulation and severe symptom induction[95].
βC1 is involved in symptom induction by interacting with plant defence machinery and suppressing TGS and PTGS. Plant infection with betasatellites damages the chloroplast structure by increasing starch accumulation, lowering thylakoid content, and altering the grana[46]. Plastoglobulins, which are involved in the protection of thylakoids, are accumulated in chloroplasts. As a result, it could be a stress response of plants to betasatellite infection[96]. C1's NLS can accumulate protein in nuclei, causing symptom development and host defence suppression[87].
The promoter's genetic elements of the βC1 of some betasatellites, like TYLCCNB, influence symptom induction by interaction with distinct host factors, inducing tissue-specific expression of target genes and playing a role in silencing the PTGS machinery and methylation-mediated plant suppression. The TYLCCNB promoter is phloem-specific and capable of producing thickening in veins in the host, whereas the tobacco curly shoot virus (TbCSV) promoter seems unable to generate similar symptoms[97]. Moreover, βC1 has an A-rich region which acts as a putative enhancer element of the gene promoter, so, that might have a role in regulating symptom severity through changes in the expression level of this gene[98].
9 UNDERSTANDING OFFENSIVE STRATEGIES FOR VIRUS MANAGEMENT
The integrated pest management (IPM) approach is a general management measure, not even for geminiviruses. IPM strategies can be used for integrated pest management, which boosts farmer revenue while enhancing productivity and the quality of finished items. The employment of such approaches not only minimises pests and diseases but also lowers the excessive use of chemicals. These measures can be taken before, during, and after crop cultivation. IPM approaches before cultivation include the use of resistance sources and virus-free planting materials, such as transplants and propagative materials, as well as the timing of planting and field placement. Resistance sources and planting materials that are virus-free are serious factors before the growing season. Introgression of resistance genes through conventional breeding methods can offer 100% protection against geminivirus infections and is the most economically and ecologically sustainable control measure[99]. These resistance genes mostly come from wild species or landraces. Numerous geminivirus resistance genes of uncultivated relatives are often used in farming, to provide effective protection against geminiviruses. Some recent studies on Solanum chilense accessions and Ty-gene have described how allelic variants of clones of these genes can be identified using virus-induced gene silencing in conjunction with allele mining. Here, they emphasise the role of different genes found in S. chilense, one of the most popular wild tomato relatives, which are obstructed by geminivirus infections. S. chilense is effectively used for introgressive hybridization to provide resistance against TYLCV, as over 80% of its accessions are impervious[100]. According to some studies, researchers found the involvement of the Rep/C1 gene in triggering the Ty-2-mediated resistance in TYLCV-IL[101]. A few studies on disease management have also revealed the importance of gene resistance during co-infection. Ty-1 to Ty-6 genes are the best examples of begomovirus resistance genes found in wild tomato species that have been introduced into many tomato cultivars with conventional breeding methods, providing high levels of resistance against TYLCV[102]. According to further research, the Ty-1 and Ty-3 genes express an RNA-dependent RNA polymerase (RDR) comparable to RDRs 3, 4, and 5 in A. thaliana, implying a function for RNA interference. Ty-2 gene encoding the protein NB-LRR is well-known for providing resistance to TYLCV[103]. Ty-4 and Ty-6 genes are also known as resistance genes, whose encoded proteins are not yet evidently explained[104]. The Ty-5 resistance gene is pelota, which expresses an mRNA surveillance element homolog[105]. Other examples are the three resistance genes cassava mosaic disease (CMD)1, CMD2 and CMD3 that confer resistance in cassava against CMD[106]. CMD2 is dominant and monogenic thus there is the possibility of viral evolution overcoming resistance[106]; In contrast, CMD1 and CMD3 are recessive, and CMD1 looks to have the highest level of resistance among the three[107].
Tomato, cotton, and cassava plants with resistance genes have effectively reduced yield losses caused by begomoviruses[107]. However, geminiviruses' potential to circumvent genetic resistance due to their rapid evolution has become a particularly difficult feature of geminivirus-plant interactions. As a result, the insertion of resistance genes obtained from wild landraces or species-acceptable economic cultivars is possible[108]. This will also make it difficult for them to realise their full potential
Growing virus-free clones of begomovirus species is critical for reducing the first inoculum and delaying the establishment of a geminivirus disease[109]. The primary source of virus dissemination is propagation materials. Virus-free mother plants are used in crops grown from cuttings (e.g., cassava) or rooted/grafted cuttings (e.g., grapevines) should be used as the source for the establishment of commercial fields. Apical meristem and tip culture are the technologies that allow for the quick generation of virus-free planting materials in a short amount of time[110].
Cultural practises as a management strategy are focused on the elimination of primary inoculums and providing incomplete interaction among viruses, susceptible hosts, and the environment, which prevent the inoculum transfer from being diseased onto healthy plants[110]. Growing crops, especially vegetables, in greenhouses with plastic or net covers and protecting them from insect vectors is also a way to reduce primary inoculums[111]. These measures will greatly reduce losses because of begomovirus infections in tropical and subtropical areas where the vector (B. tabaci) is alive during the year[112]. Planting times and field sites are the next steps to prevent the invasion of begomoviruses. Delayed sowing of annual crops whenever the insect vector numbers and inoculum supplies are at their lowest conceivable levels helps the immature plants that are extremely susceptible. Establishing the fields in areas distant from infected fields or cultivating the crops against the wind and planting barrier crops, like maize, between fields can help to reduce vector movement between them[110]. Implementation of host-free periods also reduces levels of inoculum at the start of the growing season, when plants are most susceptible to virus infection[111]. Wild plants, grown during and after the growing season, can be hosts of insect vectors and infected by species of begomoviruses as well. Removing these wild plants will disrupt their role as reservoirs or alternative hosts and reduce sources of begomoviruses’ primary inoculums.
Vector management with insecticide is the most used measure during the growing season and would be more effective when used in combination with other measures in an IPM program. Intensive cultivation and monoculture of crops year-round and greenhouse cultivation result in whitefly propagation during the year[111]. Monitoring vector populations is essential to deciding the application of insecticides[110]. Insecticides should be used when vector numbers reach a specified threshold, which must be determined for each region, because of changes in the biology of the whiteflies and the emergence of the supervector biotype B population, which is resistant to various insecticides. Since B. tabaci can effectively detoxify, metabolize, or alter toxic plant compounds and pesticides, consideration of various factors in the use of insecticides should be a focus. However, this includes the use of those insecticides that must act quickly whenever the abundance of vectors is minimal. Furthermore, insecticide rotation with different modes of action, applying broad-spectrum insecticides, and during a planting season, only using the prescribed amount and number of applications might help to break the resistance chain of the vector. Aside from the negative effects on human health and the environment, the widespread insecticide use has caused resistance in insects and the eradication of whitefly's natural enemies[10]. Biological insecticides and biological control approaches are environmentally friendly alternatives to manage B. tabaci and begomovirus diseases in protected cultures[113]. Predators, parasitoids, and fungi are the biological agents used in the reduction of whitefly populations. In addition, some botanical extracts have been known as viral inhibitors and have been used against plant viral infections[114]. These proteinaceous antiviral compounds that are endogenous viral inhibitors have been reported to reduce the infectivity of begomoviruses by 60-80% and probably induce resistance. They are extremely effective at reducing virus infectivity[115].
Geminiviruses engage in intricate yet coordinated interactions with a variety of host factors to allow and propagate effective infection cycles. Although most geminivirus-encoded proteins have numerous functions, such as altering hormone responses, manipulating protein signaling, suppressing defence responses, exploiting cell cycle regulation cascades, and hijacking ubiquitin-proteasomal pathways. Subsequently, plants have also evolved numerous multi-layered defence strategies against geminivirus infection and distribution[106]. Engineered resistance can interfere with viral proteins by acting on proteins essential for viral DNA itself or viral replication. In this regard, an "immunomodulation" mechanism was recently studied, where the transgenic plants express antibodies against viral proteins, such as anti-Rep antibody expression and single-chain antibodies generated against CP, which can provide resistance to geminiviruses[115]. Moreover, the expression of a recombinant antibody, i.e., DNase 3D8, that has single- and double-stranded non-specific DNase function, inhibited elevated amounts of BSCTV and BCTV viral DNA build-up[35]. In context, an engineered peptide sequence called "peptide aptamers" works similarly to antibodies, and they were found to disrupt the functions of TGMV and TYLCV proteins[116]. In multiple systems, transgenic approaches have been established to create plants that generate antiviral substances such as synthetic transacting siRNAs, lengthy non-coding RNAs, and artificial miRNAs with distinctive levels of success[117]. This has also been explained: resistance is determined by the sequence and only defends certain kinds of viruses. That inhibits the method's use but does not exclude its use when the prevalent virus is a distinct species[118]. CRISPR/Cas is a novel, specialised editing technology for targeting viral genes for generating resistance to several different geminiviruses[119]. During subsequent infections, a single guide RNA (sgRNA) molecule directed Cas9 endonuclease to scan invading DNA and cleave the target sequence at the protospacer-associated motif site. Cas9 recognises this trinucleotide sequence and requires it to bind to target DNA molecules[120]. SgRNAs designed to target a conserved non-nucleotide sequence among geminiviruses and betasatellites of begomoviruses could be an effective approach to reduce geminivirus infection when mixed infections occur. For instance, this approach confers multiple-virus resistance in N. benthamiana, which was infected with both TYLCV and BCTV. According to research, the level of Cas9 translation was a determining factor in the degree of symptom decrease[121].
Nevertheless, when expressed in A. thaliana, off-target Cas9 actions throughout the genome of a plant have been uncovered. SpCas9 mutant and modified gRNA scaffold (expressing the gRNA next to tRNA9met) gave fewer off-target effects[122]. On the other hand, the CRISPR/Cas9 platform could potentially be utilised to determine host characteristics that influence plant resistance and viral infection susceptibility, and to investigate the evolution of the viral genomes to find how these viruses can escape recognition by the plant immunity system[110].
Genetic engineering is the preponderance of current techniques for controlling begomovirus disease epidemics in a diverse assortment of crops produced globally. However, because of the public’s contentious view of genetically modified organisms, transgenic plants have not reached the level of commercialization. Induction of an RNA silencing response in plants has effectively produced highly resistant transgenic plants that specifically degrade or methylate the genome of the target begomovirus. The characteristics of a pathosystem are important in determining plant infection by specific viruses. There is data on geminivirus resistance factors and resistant cultivars, but it is also crucial to analyse the variables that contribute to virus infections in susceptible and resistant cultivars. Recent tests indicating novel antiviral strategies used by hosts are promising. The ability of certain geminivirus proteins to avoid host-mediated resistance via novel pathways has provided fresh insights into the development of antiviral approaches against geminiviruses. Plant tolerance can be overcome by swapping "susceptible" viral strains with recombinant or mutated "resistant" strains, or by combining infections. These situations underscore the important need for a description of mutualistic relationships and an in-depth analysis of geminiviruses and insect vectors, particularly those found in the tropics[117]. Many additional innovative and possibly powerful antiviral methods, however, require more development. Further research in a particular field may lead to the discovery of previously unknown natural resistance factors, which might provide new insight and a better understanding of host-virus interactions to continue developing more resistant crops that do not affect plant growth and development against rapid viral evolution.
10 CONCLUSION
In contrast to begomoviruses, which appear to be the exception, the majority of plant viruses are RNA viruses, showing that DNA viruses do not function as well in the subcellular machinery of plants. Understanding the interface components and mechanisms is crucial to determining how a begomovirus infects a plant. The analysis of the proteome and global gene expression of virus-infected host cells can provide in-depth knowledge on these mechanisms. Numerous studies have found that the RNA silencing apparatus, PTGS, and TGS are critical for maintaining the host's tolerance levels during begomovirus infections. But the precise mechanism is still unknown. Understanding how hosts efficiently activate silencing pathways and distribute siRNAs throughout plant tissues requires technologies like next-generation sequencing and microarrays, which may also be used to comprehend the underlying molecular pathways. Using all of this information, a crop genotype that is resistant to begomovirus infection can be developed.
Acknowledgements
RK Gaur is thankful to Uttar Pradesh Higher Education Department (UPHED, File No. 1/2022/01/04-2022-4(28)/21 dated 04/01/2022) for providing the financial support during this study.
Conflicts of Interest
The authors declared that they have no conflict of interest.
Author Contribution
Pandey V, Srivastava A, Shahmohammadi N and Nehra C wrote the manuscript. Gaur RK and Golnaraghi A were the mentors and supervised the technical aspects of the experiments and edited the manuscript. All authors read and approved the manuscript for submission.
Abbreviation List
ACMV, African cassava mosaic virus
ADK, Adenosine kinase
AS1, Asymmetric leaf 1
BAK1, Brassinosteroid insensitive 1
BCTV, Beet curly top virus
BR, Brassinosteroid
BSCTV, Beet severe curly top virus
CaLCuV, Cabbage leaf curl virus
CLCuMA, Cotton leaf curl Multan alphasatellite
CLCuMB, Leaf curl Multan Β-satellite
CLCuMV, Cotton leaf curl Multan virus
CMD, Cassava mosaic disease
CP, Coat protein
CSN, COP9 signalosome
dsDNA, Double stranded DNA
EACMCV, East African cassava mosaic virus
ETI, Effector-triggered immunity
HIR1, Hypersensitive induced reaction 1
HR, Hypersensitive response
JA, Jasmonic acid
JAZ, Jasmonate-ZIM-domain
LOX, Lipoxygenase
miRNAs, MicroRNA
MP, Movement protein
MYMV, Mungbean yellow mosaic virus
NB-LRR, Nucleotide-binding leucine-rich repeat
NIG, NSP-interacting GTPase
NLS, Nuclear localization signals
NSP, Nuclear shuttle protein
ORF, Open reading frame
PAMP, Pathogen-associated molecular pattern
PRRs, Pattern recognition receptors
PTGS, Post-transcriptional gene silencing
PTI, pattern-triggered immunity
RDR, RNA-dependent RNA polymerase
REP, Replication-associated protein
RNAi, RNA interference
SA, Salicylic acid
SCF, Skp1/Cullin1/F-box
SCTV, Spinach curly top virus
sgRNA, Single guide RNA
siRNA, Small interfering RNA
SNF1, Sucrose-nonfermenting 1
SnRK1, SNF1-related kinase
ssDNA, Single-stranded DNA
TGMV, Tomato golden mosaic virus
TGS, Transcriptional gene silencing
ToLCPalV, Tomato leaf curl Palampur virus
TYLCCNV, Tomato yellow leaf curl China virus
TYLCSV, Tomato yellow leaf curl Sardinia virus
TYLCV, Tomato yellow leaf curl virus
VSRs, Viral suppressor proteins
References
[1] Fiallo-Olivé E, Lett JM, Martin DP et al. ICTV virus taxonomy profile: Geminiviridae 2021. J Gen Virol, 2021; 102: 001696. DOI: 10.1099/jgv.0.001696
[2] Ramesh SV, Sahu PP, Prasad M et al. Geminiviruses and plant hosts: A closer examination of the molecular arms race. Viruses, 2017; 9: 256. DOI: 10.3390/v9090256
[3] Kil EJ, Byun HS, Hwang H et al. Tomato Yellow Leaf Curl Virus Infection in a Monocotyledonous Weed (Eleusine indica). Plant Pathol J, 2021; 37: 641-651. DOI: 10.5423/PPJ.FT.11.2021.0162
[4] Yazdani-Khameneh S, Golnaraghi A. The status of begomoviruses in Iran. Begomoviruses Occurrence Manag Asia Afr, 2017; 229-253. DOI: 10.1007/978-981-10-5984-1_14
[5] Zerbini FM, Briddon RW, Idris A et al. ICTV virus taxonomy profile: Geminiviridae. J Gen Virol, 2017; 98: 131-133. DOI: 10.1099/jgv.0.000738
[6] Hanley-Bowdoin L, Bejarano ER, Robertson D et al. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat Rev Microbiol, 2013; 11: 777-788. DOI: 10.1038/nrmicro3117
[7] Gafni Y, Epel BL. The role of host and viral proteins in intra- and inter-cellular trafficking of geminiviruses. Physiol Mol Plant Pathol, 2002; 60: 231-241. DOI: 10.1006/pmpp.2002.0402
[8] Lozano G, Trenado HP, Fiallo-Olivé E et al. Characterization of non-coding DNA satellites associated with sweepoviruses (genus begomovirus, geminiviridae) - Definition of a distinct class of begomovirus-associated satellites. Front Microbiol, 2016; 17: 7162. DOI: 10.3389/fmicb.2016.00162
[9] Briddon RW, Martin DP, Roumagnac P et al. Alphasatellitidae: A new family with two subfamilies for the classification of geminivirus‐ and nanovirus‐associated alphasatellites. Arch Virol, 2018; 163: 2587-2600. DOI: 10.1007/s00705-018-3854-2
[10] Idris AM, Shahid MS, Briddon RW et al. An unusual alphasatellite associated with monopartite begomoviruses attenuates symptoms and reduces betasatellite accumulation. J Gen Virol, 2011; 92: 706-717. DOI: 10.1099/vir.0.025288-0
[11] Mansourpour M, Gallet R, Abbasi A et al. Effects of an alphasatellite on the life cycle of the Nanovirus faba bean necrotic yellows virus. J Virol, 2013; 96: e0138821. DOI: 10.1128/JVI.01388-21
[12] Nawaz-ul-Rehman MS, Nahid N, Hassan M et al. Betasatellites and deltasatelliles (Tolecusatellitidae). Acad Press Encyclopedia Virol, 2021; 3: 239-246. DOI: 10.1016/B978-0-12-809633-8.21562-8
[13] Shafiq M, Qurashi F, Mushtaq S et al. DNA plant viruses: biochemistry, replication, and molecular genetics. Appl Plant Virol, 2020; 169-182. DOI: 10.1016/B978-0-12-818654-1.00013-X
[14] Ferro CG, Zerbini FM, Navas-Castillo J et al. Revealing the complexity of sweepovirus-deltasatellite-plant host interactions: Expanded natural and experimental helper virus range and effect dependence on the virus-host combination. Microorg, 2021; 9: 1018. DOI: 10.3390/microorganisms9051018
[15] Fiallo‐Olivé E, Tovar R, Navas‐Castillo J. Deciphering the biology of deltasatellites from the New World: Maintenance by New World begomoviruses and whitefly transmission. New Phytol, 2016; 212: 680-692. DOI: 10.1111/nph.14071
[16] Ghanim M, Morin S, Czosnek H. Rate of tomato yellow leaf curl virus translocation in the circulative transmission pathway of its vector, the whitefly Bemisia tabaci. Phytopathol, 2018; 91: 188-196. DOI: 10.1094/PHYTO.2001.91.2.188
[17] Rana VS, Singh ST, Priya NG et al. Arsenophonus GroEL interacts with CLCuV and is localized in the midgut and salivary gland of whitefly B. tabaci. PLoS One, 2012; 7: e42168. DOI: 10.1371/journal.pone.0042168
[18] Cathrin PB, Ghanim M. Recent advances on interactions between the whitefly Bemisia tabaci and begomoviruses, with emphasis on Tomato yellow leaf curl virus. Plant Virus Host Interact, 2014; 79-103. DOI: 10.1016/B978-0-12-411584-2.00004-4
[19] Pan H, Chu D, Yan W et al. Rapid spread of Tomato yellow leaf curl virus in China is aided differentially by two invasive whiteflies. PloS One, 2012; 7: e34817. DOI: 10.1371/journal.pone.0034817
[20] Gilbertson RL, Batuman O, Webster CG et al. Role of insect super vectors Bemisia tabaci and Frankliniella occidentalis in plant virus emergence and global spread. Annu Rev Virol, 2015; 2: 67-93. DOI: 10.1146/annurev-virology-031413-085410
[21] Bruce G, Gu M, Shi N et al. Influence of retinoblastoma-related gene silencing on the initiation of DNA replication by African cassava mosaic virus Rep in cells of mature leaves in Nicotiana benthamiana plants. Virol J, 2011; 8: 561. DOI: 10.1186/1743-422X-8-561
[22] Hanley-Bowdoin L, Bejarano ER, Robertson D et al. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol, 2013; 11: 777-788. DOI: 10.1038/nrmicro3117
[23] Suyal G, Rana VS, Mukherjee SK et al. Arabidopsis thaliana NAC083 protein interacts with Mungbean yellow mosaic India virus (MYMIV) Rep protein. Virus Genes, 2014; 48: 486-493. DOI: 10.1007/s11262-013-1028-6
[24] Florentino LH, Santos AA, Fontenelle MR et al. A PERK-like receptor kinase interacts with the geminivirus nuclear shuttle protein and potentiates viral infection. J Virol, 2006; 80: 6648-6656. DOI: 10.1128/JVI.00173-06
[25] Dogra SC, Eini O, Rezaian MA et al. A novel shaggy-like kinase interacts with the Tomato leaf curl virus pathogenicity determinant C4 protein. Plant Mol Biol, 2009; 71: 25-38. DOI: 10.1007/s11103-009-9506-x
[26] Wang MB, Masuta C, Smith NA et al. RNA silencing and plant viral diseases. Mol Plant Microbe Interact, 2012; 25: 1275-1285. DOI: 10.1094/MPMI-04-12-0093-CR
[27] Egelkrout EM, Robertson D, Hanley-Bowdoin L. Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell, 2001; 13: 1437-1452. DOI: 10.1105/tpc.13.6.1437
[28] Tena G, Boudsocq M, Sheen J. Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol, 2011; 14: 519-529. DOI: 10.1016/j.pbi.2011.05.006
[29] Lacatus G, Sunter G. The Arabidopsis PEAPOD2 transcription factor interacts with geminivirus AL2 protein and the coat protein promoter. Virol, 2009; 392: 196-202. DOI: 10.1016/j.virol.2009.07.004
[30] Li H, Zeng R, Chen Z et al. S-acylation of a geminivirus C4 protein is essential for regulating the CLAVATA pathway in symptom determination. J Exp Bot, 2018; 69: 4459-4468. DOI: 10.1093/jxb/ery228
[31] Zhou Y, Xu Z, Duan C et al. Dual transcriptome analysis reveals insights into the response to Rice black-streaked dwarf virus in maize. J Exp Bot, 2016; 67: 4593-4609. DOI: 10.1093/jxb/erw244
[32] Kaur H, Yadav CB, Alatar AA et al. Gene expression changes in tomato during symptom development in response to leaf curl virus infection. J Plant Biochem Biotechnol, 2014; 24: 347-354. DOI: 10.1007/s13562-014-0280-8
[33] Sun X, Wang Z, Gu Q et al. Transcriptome analysis of Cucumis sativus infected by Cucurbit chlorotic yellows virus. Virol J, 2017; 14: 18. DOI: 10.1186/s12985-017-0690-z
[34] Nuruzzaman M, Sharoni AM, Satoh K et al. NAC transcription factor family genes are differentially expressed in rice during infections with rice dwarf virus, rice black-streaked dwarf virus, rice grassy stunt virus, rice ragged stunt virus, and rice transitory yellowing virus. Front Plant Sci, 2015; 6: 676. DOI: 10.3389/fpls.2015.00676
[35] Lee G, Shim HK, Kwon MH et al. A nucleic acid hydrolyzing recombinant antibody confers resistance to curtovirus infection in tobacco. Plant Cell Tissue Organ Cult, 2013; 115: 179-187. DOI: 10.1007/s11240-013-0357-4
[36] Mishra GP, Singh B, Seth T et al. Biotechnological advancements and begomovirus management in okra (Abelmoschus esculentus L.): Status and perspectives. Front Plant Sci, 2017; 8: 360. DOI: 10.3389/fpls.2017.00360
[37] Pusztahelyi T, Holb IJ, Pócsi I. Plant-fungal interactions: special secondary metabolites of the biotrophic, necrotrophic, and other specific interactions. Fungal Metab, 2017; 133-190. DOI: 10.1007/978-3-319-19456-1_39-1
[38] Rodrigo G, Carrera J, Ruiz-Ferrer V et al. A meta-analysis reveals the commonalities and differences in Arabidopsis thaliana response to different viral pathogens. PLoS One, 2012; 7: e40526. DOI: 10.1371/journal.pone.0040526
[39] Alvarado VY, Scholthof HB. AGO2: A new OsArgonaute compromising plant virus accumulation. Front Plant Sci, 2012; 2: 112. DOI: 10.3389/fpls.2011.00112
[40] Dasgupta U, Mishra GP, Dikshit HK et al. Comparative RNA-Seq analysis unfolds a complex regulatory network imparting yellow mosaic disease resistance in mungbean [Vigna radiate (L.) R. Wilczek]. PLoS One, 2021; 16: e0244593. DOI: 10.1371/journal.pone.0244593
[41] Yarunin A, Panse VG, Petfalski E et al. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J, 2005; 24: 580-588. DOI: 10.1038/sj.emboj.7600540
[42] Yang JY, Iwasaki M, Machida C. βC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev, 2008; 22: 2564-2577. DOI: 10.1101/gad.1682208
[43] Hao L, Wang H, Sunter G et al. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell, 2003; 15: 1034-1048. DOI: 10.1105/tpc.009530
[44] Chinchilla D, Zipfel C, Robatzek S et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature, 2007; 448: 497-500. DOI: 10.1038/nature05999
[45] Panas MD, Schulte T, Thaa B et al. Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathog, 2015; 11: e1004659. DOI: 10.1371/journal.ppat.1004659
[46] Mubin M, Ijaz S, Nahid N et al. Journey of begomovirus betasatellite molecules: From satellites to indispensable partners. Virus Genes, 2020; 56: 16-26. DOI: 10.1007/s11262-019-01716-5
[47] Eini O, Dogra S, Selth LA et al. Interaction with a host ubiquitin-conjugating enzyme is required for the pathogenicity of a geminiviral DNA β satellite. Mol Plant Microbe Interact, 2009; 22: 737-746. DOI: 10.1094/MPMI-22-6-0737
[48] Verhage A, van Wees SC, Pieterse CM. Plant immunity: It is the hormones talking, but what do they say? Plant Physiol, 2010; 154: 536-540. DOI: 10.1104/pp.110.161570
[49] Bosamia TC, Dodia SM, Mishra GP et al. Unraveling the mechanisms of resistance to Sclerotium rolfsii in peanut (Arachis hypogaea L.) using comparative RNA-Seq analysis of resistant and susceptible genotypes. PLoS One, 2020; 15: e0236823. DOI: 10.1371/journal.pone.0236823
[50] Rubio-Somoza I, Cuperus JT, Weigel D et al. Regulation and functional specialization of small RNA-target nodes during plant development. Curr Opin Plant Biol, 2009; 12: 622-627. DOI: 10.1016/j.pbi.2009.07.003
[51] Sahu PP, Sharma N, Puranik S. Involvement of host regulatory pathways during geminivirus infection: A novel platform for generating durable resistance. Func Integr Genom, 2017; 14: 47-58. DOI: 10.1007/s10142-013-0346-z
[52] Lozano-Durán R, Rosas-Díaz T, Gusmaroli G et al. Geminiviruses subvert ubiquitination by altering CSNmediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell, 2011; 23: 1014-1032. DOI: 10.1105/tpc.110.080267
[53] Zhang Z, Chen H, Huang X et al. BSCTV C2 attenuates the degradation of SAMDC 1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell, 2011; 23: 273-288. DOI: 10.1105/tpc.110.081695
[54] Czosnek H, Eybishtz A, Sade D et al. Discovering host genes involved in the infection by the tomato yellow leaf curl virus complex and in the establishment of resistance to the virus using tobacco rattle virus-based post transcriptional gene silencing. Viruses, 2013; 5: 998-1022. DOI: 10.3390/v5030998
[55] Schoelz JE, Harries PA, Nelson RS. Intracellular transport of plant viruses: Finding the door out of the cell. Mol Plant, 2011; 4: 813-831. DOI: 10.1093/mp/ssr070
[56] Miozzi L, Napoli C, Sardo L et al. Transcriptomics of the interaction between the monopartite phloem-limited geminivirus Tomato Yellow Leaf Curl Sardinia Virus and Solanum lycopersicum highlights a role for plant hormones, autophagy and plant immune system fine tuning during Infection. PLoS One, 2014; 9: e89951. DOI: 10.1371/journal.pone.0089951
[57] Baliji S, Lacatus G, Sunter G. The interaction between geminivirus pathogenicity proteins and adenosine kinase leads to increased expression of primary cytokinin-responsive genes. Virol, 2010; 402: 238-247. DOI: 10.1016/j.virol.2010.03.023
[58] Lozano-Duran R, Rosas-Diaz T, Luna AP et al. Identification of host genes involved in geminivirus infection using a reverse genetics approach. PLoS One, 2011; 6: e22383. DOI: 10.1371/journal.pone.0022383
[59] Luan JB, Yao DM, Zhang T et al. Suppression of terpenoid synthesis in plants by a virus promotes its mutualism with vectors. Ecol Lett, 2013; 16: 390-398. DOI: 10.1111/ele.12055
[60] Li P, Liu C, Deng WH et al. Plant begomoviruses subvert ubiquitination to suppress plant defenses against insect vectors. PLoS Pathog, 2019; 15: e1007607. DOI: 10.1371/journal.ppat.1007607
[61] Carvalho CM, Machado JPB, Zerbini FM et al. NSP-interacting GTPase: A cytosolic protein as cofactor for nuclear shuttle proteins. Plant Signal Behav, 2008; 3: 752-754. DOI: 10.1111/j.1365-313X.2008.03556.x
[62] Poornima Priyadarshini CG, Savithri HS. Kinetics of interaction of Cotton Leaf Curl Kokhran Virus-Dabawali (CLCuKV-Dab) coat protein and its mutants with ssDNA. Virol, 2009; 386: 427-437. DOI: 10.1016/j.virol.2009.01.016
[63] Martins LG, Raimundo GA, Ribeiro NG et al. A begomovirus nuclear shuttle protein-interacting immune hub: hijacking host transport activities and suppressing incompatible functions. Front Plant Sci, 2020; 11: 398. DOI: 10.3389/fpls.2020.00398
[64] Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell, 2007; 130: 413-426. DOI: 10.1016/j.cell.2007.07.039
[65] Buchmann RC, Asad S, Wolf JN et al. Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J Virol, 2009; 83: 5005-5013. DOI: 10.1128/JVI.01771-08
[66] Li F, Xu X, Huang C et al. The AC5 protein encoded by Mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. New Phytol, 2015; 208: 555-569. DOI: 10.1111/nph.13473
[67] Zhang J, Dong J, Xu Y et al. V2 protein encoded by Tomato yellow leaf curl China virus is an RNA silencing suppressor. Virus Res, 2012; 163: 51-58. DOI: 10.1016/j.virusres.2011.08.009
[68] Amin I, Hussain K, Akbergenov R et al. Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-beta-satellite complex. Mol Plant Microbe Interact, 2011; 24: 973-983. DOI: 10.1094/MPMI-01-11-0001
[69] Zrachya A, Glick E, Levy Y et al. Suppressor of RNA silencing encoded by Tomato yellow leaf curl virus-Israel. Virol, 2007; 358: 159-165. DOI: 10.1016/j.virol.2006.08.016
[70] Calil IP, Fontes EPB. Plant immunity against viruses: antiviral immune receptors in focus. Ann Bot, 2017; 119: 711-723. DOI: 10.1093/aob/mcw200
[71] Teixeira RM, Ferreira MA, Raimundo GA et al. Geminiviral triggers and suppressors of plant antiviral immunity. Microorg, 2021; 9: 775. DOI: 10.3390/microorganisms9040775
[72] Zipfel C. Plant pattern-recognition receptors. Trends Immunol, 2014; 35: 345-351. DOI: 10.1016/j.it.2014.05.004
[73] Böhm H, Albert I, Fan L et al. Immune receptor complexes at the plant cell surface. Curr Opin Plant Biol, 2014; 20: 7-54. DOI: 10.1016/j.pbi.2014.04.007
[74] Soitamo AJ, Jada B, Lehto K. Expression of geminiviral AC2 RNA silencing suppressor changes sugar and jasmonate responsive gene expression in transgenic tobacco plants. BMC Plant Biol, 2012; 12: 204. DOI: 10.1186/1471-2229-12-204
[75] Zhu JY, Sae-Seaw J, Wang ZY. Brassinosteroid signaling. Dev, 2013; 140: 1615-1620. DOI: 10.1242/dev.060590
[76] Mills LK, Deom CM. Geminivirus C4 protein alters Arabidopsis development. Protoplasma, 2010; 239: 95-110. DOI: 10.1007/s00709-009-0086-z
[77] Machado JP, Brustolini OJ, Mendes GC et al. NIK1, a host factor specialized in antiviral defense or a novel general regulator of plant immunity? Bioessays, 2015; 37: 1236-1242. DOI: 10.1002/bies.201500066
[78] Machado JPB, Calil IP, Santos AA et al. Translational control in plant antiviral immunity. Genet Mol Biol, 2017; 40: 292-304. DOI: 10.1590/1678-4685-gmb-2016-0092
[79] Gouveia BC, Calil IP, Machado JPB et al. Immune receptors and co-receptors in antiviral innate immunity in plants. Front Microbiol, 2017; 7: 2139. DOI: 10.3389/fmicb.2016.02139
[80] Brustolini OJ, Machado JPB, Condori‐Apfata JA et al. Sustained NIK mediated antiviral signaling confers broad-spectrum tolerance to begomoviruses in cultivated plants. Plant Biotechnol J, 2015; 13: 1300-1311. DOI: 10.1111/pbi.12349
[81] Santos AA, Lopes KV, Apfata JA et al. NSP interacting kinase, NIK: A transducer of plant defence signaling. J Exp Bot, 2016; 61: 3839-3845. DOI: 10.1093/jxb/erq219
[82] Li B, Ferreira MA, Huang M et al. The receptor-like kinase NIK1 targets FLS2/BAK1 immune complex and inversely modulates antiviral and antibacterial immunity. Nat Commun, 2019; 10: 4996. DOI: 10.1038/s41467-019-12847-6
[83] Krapp S, Greiner E, Amin B et al. The stress granule component G3BP is a novel interaction partner for the nuclear shuttle proteins of the nanovirus pea necrotic yellow dwarf virus and geminivirus abutilon mosaic virus. Virus Res, 2017; 227: 6-14. DOI: 10.1016/j.virusres.2016.09.021
[84] Ye J, Yang J, Sun Y et al. Geminivirus activates asymmetric leaves 2 to accelerate cytoplasmic DCP2-mediated mRNA turnover and weakens RNA silencing in Arabidopsis. PLoS Pathog, 2015; 11: e1005196. DOI: 10.1371/journal.ppat.1005196
[85] Mei Y, Ma Z, Wang Y et al. Geminivirus C4 antagonizes the HIR1-mediated hypersensitive response by inhibiting the HIR1 self-interaction and promoting degradation of the protein. New Phytol, 2020; 225: 1311-1326. DOI: 10.1111/nph.16208
[86] Jia Q, Liu N, Xie K et al. CLCuMuB βc1 subverts ubiquitination by interacting with NbSKP1s to enhance geminivirus infection in Nicotiana benthamiana. PLoS Pathog, 2016; 12: e1005668. DOI: 10.1371/journal.ppat.1005668
[87] Nair A, Chatterjee KS, Jha V. Stability of begomoviral pathogenicity determinant βC1 is modulated by mutually antagonistic SUMOylation and SIM interactions. BMC Biol, 2020; 18: 110. DOI: 10.1186/s12915-020-00843-y
[88] Teng K, Chen H, Lai J et al. Involvement of C4 protein of beet severe curly top virus (family Geminiviridae) in virus movement. PLoS One, 2010; 5: e11280. DOI: 10.1371/journal.pone.0011280
[89] Kulshreshtha A, Kumar Y, Roshan P et al. AC4 protein of tomato leaf curl Palampur virus is an RNA silencing suppressor and a pathogenicity determinant. Microb Pathog, 2019; 135: 103636. DOI: 10.1016/j.micpath.2019.103636
[90] Fondong VN. Geminivirus protein structure and function. Mol Plant Pathol, 2013; 14: 635-649. DOI: 10.1111/mpp.12032
[91] Xie Y, Zhao L, Jiao X et al. A recombinant begomovirus resulting from exchange of the C4 gene. J Gen Virol, 2013; 94: 1896-1907. DOI: 10.1099/vir.0.053181-0
[92] Mei Y, Wang Y, Hu T et al. Nucleocytoplasmic shuttling of geminivirus C4 protein mediated by phosphorylation and myristoylation is critical for viral pathogenicity. Mol Plant, 2018; 11: 1466-1481. DOI: 10.1016/j.molp.2018.10.004
[93] Mei Y, Yang X, Huang C et al. Tomato leaf curl Yunnan virus-encoded C4 induces cell division through enhancing stability of Cyclin D 1.1 via impairing NbSKη-mediated phosphorylation in Nicotiana benthamiana. PLoS Pathog, 2018; 14: e1006789. DOI: 10.1371/journal.ppat.1006789
[94] Gong P, Tan H, Zhao S et al. Geminiviruses encode additional small proteins with specific subcellular localizations and virulence function. Nat Commun, 2021; 12: 4278. DOI: 10.1038/s41467-021-24617-4
[95] Zhong X, Wang ZQ, Xiao R et al. iTRAQ analysis of the tobacco leaf proteome reveals that RNA directed DNA methylation (RdDM) has important roles in defense against geminivirus-betasatellite infection. J Proteom, 2017; 152: 88-101. DOI: 10.1016/j.jprot.2016.10.015
[96] Bhattacharyya D, Gnanasekaran P, Kumar RK et al. A geminivirus betasatellite damages the structural and functional integrity of chloroplasts leading to symptom formation and inhibition of photosynthesis. J Exp Bot, 2015; 66: 5881-5895. DOI: 10.1093/jxb/erv299
[97] Ding C, Qing L, Li Z et al. Genetic determinants of symptoms on viral DNA satellites. Appl Environ Microbiol, 2009; 75: 5380-5389. DOI: 10.1128/AEM.01193-09
[98] Tao X, Qing L, Zhou X. Function of A-Rich region in DNAβ associated with tomato yellow leaf curl China virus. Chin Sci Bull, 2004; 49: 1490-1493. DOI: 10.1360/04wc0132
[99] Ariyo OA, Atiri GI, Dixon AGO et al. The use of biolistic inoculation of cassava mosaic begomoviruses in screening cassava for resistance to cassava diseases. J Virol Methods, 2006; 137: 43-50. DOI: 10.1016/j.jviromet.2006.05.031
[100] Yan Z, Pérez-de-Castro A, Díez MJ et al. Resistance to Tomato yellow leaf curl virus in tomato germplasm. Front Plant Sci, 2018; 9: 1198. DOI: 10.3389/fpls.2018.01198
[101] Shen X, Yan Z, Wang X et al. The NLR protein encoded by the resistance gene Ty-2 is triggered by the replication-associated protein Rep/C1 of tomato yellow leaf curl virus. Front Plant Sci, 2020; 11: 1-17. DOI: 10.3389/fpls.2020.545306
[102] Caro M, Verlaan MG, Julián O et al. Assessing the genetic variation of Ty-1 and Ty-3 alleles conferring resistance to Tomato yellow leaf curl virus in a broad tomato germplasm. Mol Breed, 2015; 35: 132. DOI: 10.1007/s11032-015-0329-y
[103] Yamaguchi H, Ohnishi J, Saito A et al. An NB-LRR gene, TYNBS1, Is responsible for resistance mediated by the Ty-2 begomovirus resistance locus of tomato. Theor Appl Genet, 2018; 131: 1345-1362. DOI: 10.1007/s00122-018-3082-x
[104] Scott JW, Hutton SF, Freeman JH. Fla. 8638B and Fla. 8624 tomato breeding lines with begomovirus resistance genes ty-5 plus Ty-6 and Ty-6, respectively. Hort Sci, 2015; 50: 1405-1407. DOI: 10.21273/HORTSCI.50.9.1405
[105] Lapidot M, Karniel U, Gelbart D. A novel route controlling begomovirus resistance by the messenger RNA surveillance factor Pelota. PLoS Genet, 2015; 11: e1005538. DOI: 10.1371/journal.pgen.1005538
[106] Beam K, Ascencio-Ibañez JT. Geminivirus resistance: A minireview. Front Plant Sci, 2020; 11: 1131. DOI: 10.3389/fpls.2020.01131
[107] Okogbenin E, Egesi CN, Olasanmi B et al. Molecular marker analysis and validation of resistance to cassava mosaic disease in elite cassava genotypes in Nigeria. Crop Sci, 2012; 52: 2576-2586. DOI: 10.2135/cropsci2011.11.0586
[108] Kuria P, Ilyas M, Ateka E et al. Differential response of cassava genotypes to infection by cassava mosaic geminiviruses. Virus Res, 2017; 227: 69-81. DOI: 10.1016/j.virusres.2016.09.022
[109] de Ronde D, Butterbach P, Kormelink R. Dominant resistance against plant viruses. Front Plant Sci, 2014; 5: 307. DOI: 10.3389/fpls.2014.00307
[110] Rojas MR, Macedo MA, Maliano MR et al. World management of geminiviruses. Annu Rev Phytopathol, 2018; 56: 637-677. DOI: 10.1146/annurev-phyto-080615-100327
[111] Saeed ST, Samad A. Emerging threats of begomoviruses to the cultivation of medicinal and aromatic crops and their management strategies. Virus Dis, 2017; 28: 1-17. DOI: 10.1007/s13337-016-0358-0
[112] Ning W, Shi X, Liu B et al. Transmission of Tomato yellow leaf curl virus by Bemisia tabaci as affected by whitefly sex and biotype. Sci Rep, 2015; 5: e10744. DOI: 10.1038/srep10744
[113] Stansly PA, Sánchez PA, Rodrıguez JM et al. Prospects for biological control of Bemisia tabaci (Homoptera, Aleyrodidae) in greenhouse tomatoes of southern Spain. Crop Prot, 2004; 23: 701-712. DOI: 10.1016/j.cropro.2003.11.016
[114] Safarzadeh-Khosroshahi T, Rakhshandehroo F, Larijani K et al. Study on the antiviral activity of yarrow ethanolic extract against Cucumber mosaic virus. The 1st International & 13th Iranian Genetic Congress, Tehran, Iran, May 17, 2014.
[115] Zakri AM, Ziegler A, Commandeur U et al. In vivo expression and binding activity of ScFv-RWAV, which recognizes the coat protein of tomato leaf curl new delhi virus (Family Geminiviridae). Arch Virol, 2012; 157: 1291-1299. DOI: 10.1007/s00705-012-1310-2
[116] Reyes MI, Nash TE, Dallas MM et al. Peptide aptamers that bind to geminivirus replication proteins confer a resistance phenotype to Tomato yellow leaf curl virus and Tomato mottle virus infection in tomato. J Virol, 2013; 87: 9691-9706. DOI: 10.1128/JVI.01095-13
[117] Kumar A, Khan JA. Geminivirus Resistance Strategies. Springer Nature Switzerland Ag: Handel, Switzerland, 2019. DOI: 10.1007/978-3-030-18248-9_11
[118] Fuentes A, Carlos N, Ruiz Y et al. Field trial and molecular characterization of RNAi-transgenic tomato plants that exhibit resistance to tomato yellow leaf curl geminivirus. Mol Plant Microbe Interact, 2016; 29: 197-209. DOI: 10.1094/MPMI-08-15-0181-R
[119] Ji X, Si X, Zhang Y et al. Conferring DNA virus resistance with high specificity in plants using the virus-inducible genome-editing system. Genome Biol, 2018; 19: 197. DOI: 10.1186/s13059-018-1580-4
[120] Minoia S, Petrozza A, D'Onofrio O et al. A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res Notes, 2010; 3: 69. DOI: 10.1186/1756-0500-3-69
[121] Ji X, Zhang H, Zhang Y et al. Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in Plants. Nat Plant, 2015; 1: 15144. DOI: 10.1038/nplants.2015.144
[122] Zhang Q, Xing HL, Wang ZP et al. Potential high-frequency off-target mutagenesis induced by CRISPR/Cas9 in arabidopsis and its prevention. Plant Mol Biol, 2018; 96: 445-456. DOI: 10.1007/s11103-018-0709-x
Copyright © 2023 The Author(s). This open-access article is licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, sharing, adaptation, distribution, and reproduction in any medium, provided the original work is properly cited.

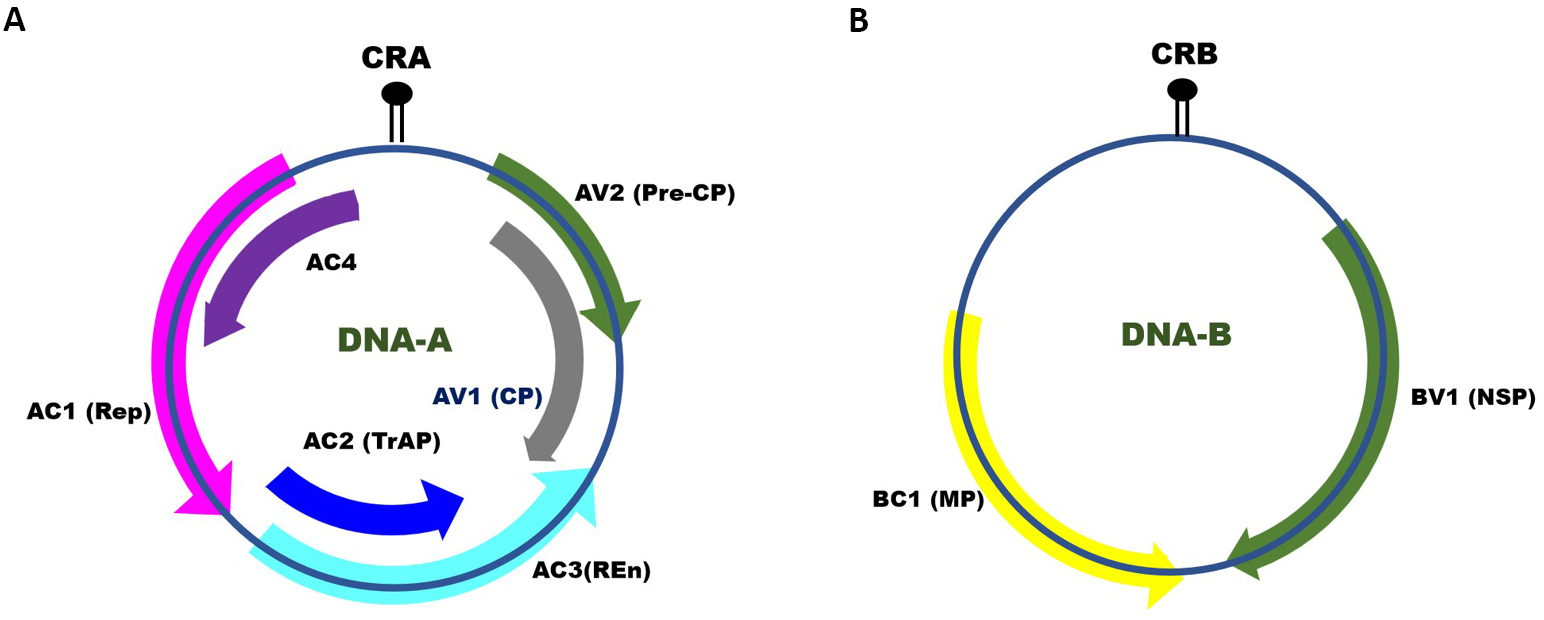
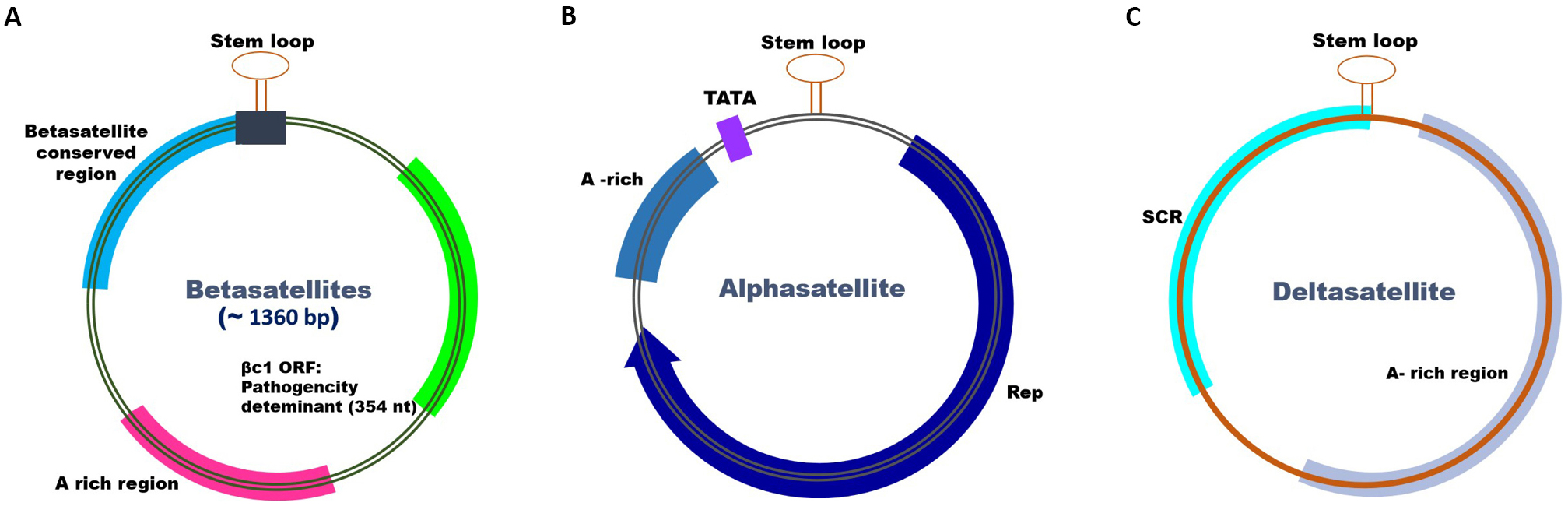
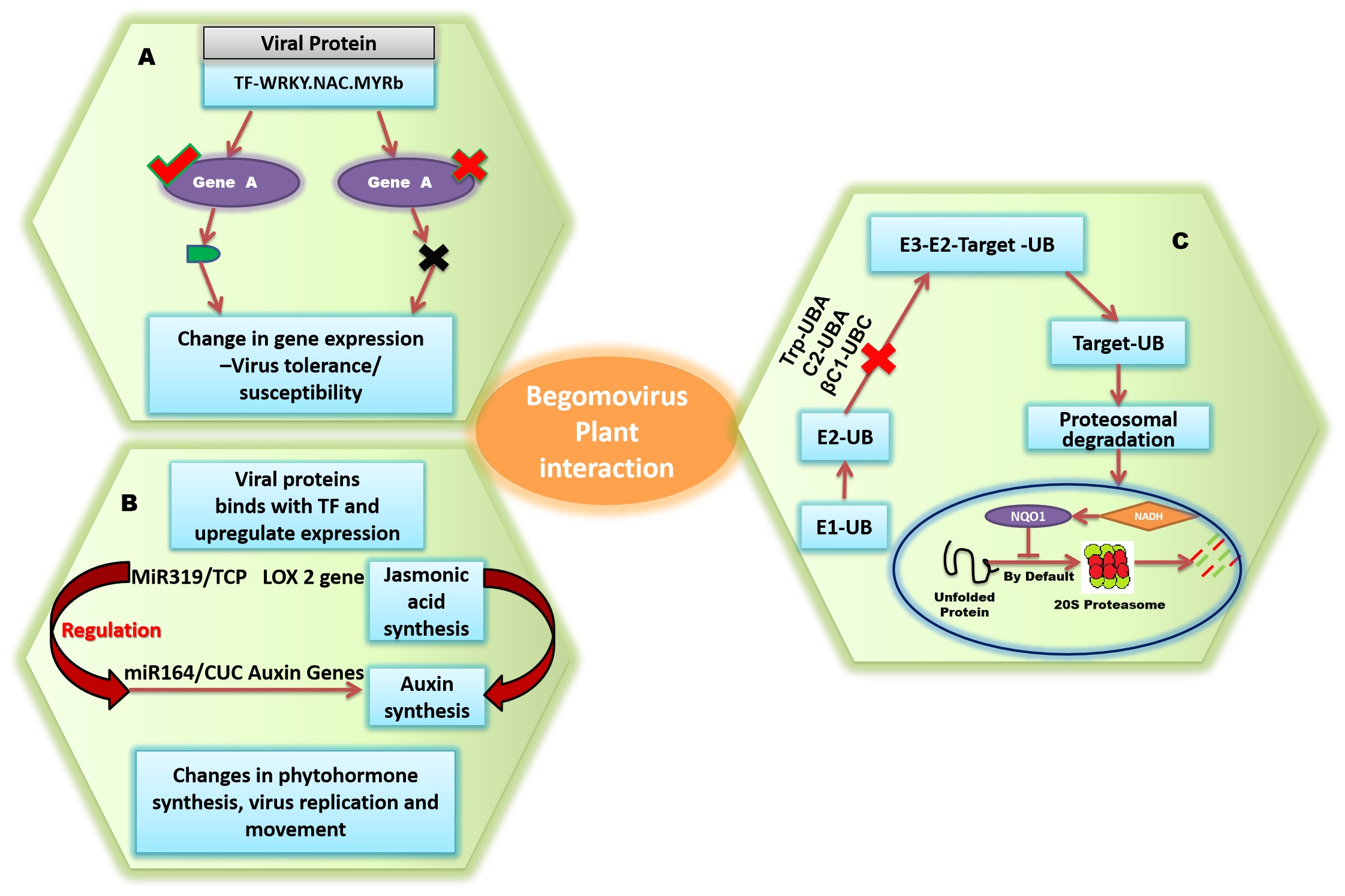




 Copyright ©
Copyright ©