A Review on Bioremediation - An Emerging Technology for Treatment of Radionuclide Waste
Humma Akram Cheema1,2*
1Department of Chemistry, University of Education, Lahore, Pakistan
2Department of Chemistry, University of Agriculture, Faisalabad, Pakistan
*Correspondence to: Humma Akram Cheema, PhD, Researcher, Department of Chemistry, University of Education Lahore, College Road, Township Block C Phase 1 Johar Town, Lahore, Punjab 54770, Pakistan or Department of Chemistry, University of Agriculture, Jail Road, Al-Khidmat Police Markaz Police Lines, Faisalabad, Punjab 38000, Pakistan; Email: humcheema242@yahoo.com
Abstract
Radioactive nuclides are utilized in large quantities in different sectors such as nuclear power plants to address energy demand. As a result, there is a massive amount of radioactive waste produced from nuclear sites and its spent sources including mining activities, nuclear weapons recycling, nuclear weapon, and nuclear energy generation. The widespread discharge of radionuclides into the environment and their mobility is a worldwide matter of concern given its health risks to living organisms. Hence, there is a need to embrace sustainable technologies for the effective management of radioactive nuclide wastes. Bioremediation is a promising and eco-friendly approach to remediate radioactive waste. The current review discussed various modes of bioremediation such as microbial, phyto, and myco-remediation for the management of radioactive waste. The interaction of microorganisms with various actinides and their fission products are also discussed. Moreover, a sustainable and efficient strategy for radioactive nuclide waste management in developing countries is also highlighted.
Keywords: radioactive wastes, bioremediation, actinides fission products, radioactive waste management
1 INTRODUCTION
Atoms with sufficient nuclear energy to destabilize them are called radionuclides. This excessive nuclear energy can be released in three different ways including alpha, beta and gamma radiation[1]. The emission of excessive nuclear energy from nucleus in forms of alpha, beta and gamma radiation is known as radioactive decay/radioactivity[2]. Both natural and anthropogenic sources originate radiation in the environment. It is estimated that more than 80% of the worldwide environmental radiation is from natural sources[3]. Naturally occurring radionuclide can be grouped into three types viz. primordial, secondary radionuclides and cosmogenic radionuclides. Primary radionuclides produce mainly from the interiors of stars as a part of the earth crust, while secondary radionuclides are derived from the decay of primary radionuclides and are radiogenic isotopes. Primordial radionuclides have longer half-lives in contrast to secondary radionuclides. Cosmic radionuclides are generated after interactions of cosmic rays[4,5].
It is commonly believed that radionuclides can be generated anthropogenically by nuclear reactors, particle accelerators, or radionuclide generators. They are continuously released into the environment following nuclear power weapon tests, nuclear energy activities, and nuclear power plant accidents[6-9]. In the previous six decades, the list of generated radioactive nuclide by fission reactors is constantly elongating and includes Neptunium-237, Plutonium-238, Americium-241, Hydrogen-3, Carbon-14, Krypton-85, Strontium-90, Technetium-99, Iodine-129 and Cesium-137 along uranium from different nuclear sites[10]. Radionuclides have been found in water, soil and air currents around the globe, and they are usually dumped on the ground depending upon their weight. At times, heavy rains may carry the radioactive elements to the ground[4]. Radioactive nuclide that persists in the soil can dissolve into solution, form complexes with organic matter in the soil, or precipitate out easily. The stasis of these radioactive nuclides in the uppermost layer of the soil is considered detrimental to environment. Radioactive nuclides are found either in soil or sediments in aquatic system where they can be easily incorporated into the food chain prompting health risks[6,11-14].
Table 1 describes the release of radioactive nuclide waste from nuclear sites and its spent sources including mining activities, industrial activities, land fillings, agrochemical waste, nuclear weapons recycling, nuclear weapon and nuclear energy generation, resulting in severe environmental hazards that cause serious health issue[15-18]. The discharge of radioactive nuclides to the environment continuously has been increased as nuclear energy demands increases day by day. As a result, the adverse and harmful effects of radionuclide waste on the environment and organisms are increasing. Table 2 summarizes the effect of various radioactive nuclide on biota along with emitted radiation. Hereafter, there is a need to implant an efficient method and strategy for the better management and treatment of radioactive nuclide waste. Currently, the removal of radioactive nuclide from all of the waste streams is a worldwide matter of concern.
Table 1. Various Streams of Radioactive Material Pollution and Various Bioremediation Modes to Manage Pollution[15-17]
Sources |
Various Steam of Radioactive Material Pollution |
|
Industrial activities |
Nuclear weapon recycling, plastics, preservatives, picroelectronics and refineries |
|
Mining activities |
Smelting, mine spoil, tailing and metal industries |
|
Waste disposal |
Sewage, sludge and leachate from landfills |
|
Agrochemicals |
Extensive use of fertilizer and pesticides |
|
Various Bio-remediation Modes |
||
Bacterial/Fungi |
Plant |
|
Bio-reduction |
Phyto-extraction |
|
Bio-accumulation |
Phyto-stabilization |
|
Bio-mineralization |
Rhizo-filtration |
|
Bio-sorption |
Phyto-volatilization |
|
Table 2. Various Radioactive Nuclide with Emitted Radiation and Effect on Biota[12-14]
Radioactive Nuclide |
Half-life |
Emitted Radiation (MeV) |
Effect on Biota |
U238, 235, 234, 239 |
69y-109y |
α, β (4.2-205) |
Dangerous and abundant, contains natural fissile elements as reactant. Absorbed by most cells/organisms |
Pu238, 239, 240, 242 |
88y-105y |
α (0.1-560) |
Highly dangerous and accumulates especially in bones. Highly fissile /reactive, sustains chain reaction |
Ra226, 228, 223, 224 |
11d-103y |
α, β (4.9-6.0) |
All isotopes are highly radioactive /dangerous. A heavy metal which incorporates easily into cells/tissues |
Rn210, 222, 224 |
2.4h-3.8d |
α, β (2.8-6.0) |
Noble gas that has only radioisotopes present. Intensely radioactive and dangerous even from natural sources |
Cs134, 137, 133 |
30y-106y |
β (1.2-2.1) |
Biological properties like K, can substitute for K in cells. Highly toxic when released into the environment |
Sr89, 90, 86, 88 |
50d-29y |
β, γ (0.6-1.2) |
Properties like Ca and can substitute for Ca in tissues. Often toxic when released into the environment |
Np237, 236, 235 |
2.5d-106y |
α, β (1.0-5.2) |
All forms are radioactive. Product of U fission, present in w |
Am241, 243, 242 |
103y-104y |
α (5.2-5.6) |
Product of U fission and often present in high levels in industrial waste. Adheres strongly to soil particles |
Tc99, 98, 97 |
4d-106y |
β, γ (0.2-0.8) |
All forms are radioactive. Product of U fission, present in waste and dangerous, used in cancer treatment |
Po208, 209, 210 |
2.8y-138y |
α, β (1.4-5.2) |
Highly dangerous, is taken-up with no biological function. |
Different physio-chemical methods have been introduced for their recovery from waste streams such as liquid-liquid extraction, chemical precipitation, electrochemical processes, coagulation, ion exchange, membrane processes and co-precipitation[19]. However, these methods have some drawbacks such as their low cost-effectiveness, high degree of selectivity, and the need for optimal environmental conditions to improve their efficiency. Therefore, this review focuses on “Bioremediation” for the recovery of radioactive nuclides from spent nuclear sources. It is a process that based on metabolic activity of microbes for the removal of radioactive nuclide from the waste. Due to the commercial benefits of microbial systems such as their low costs and easy cultivation without chemical contaminations produced during the biological process, the microbial system has been adapted worldwide for the efficient removal of radioactive waste nuclides[20]. Therefore, biotechnology performs a significant role in different industrial sectors in favor of environmental protection[15]. Different research programs have been introduced to access the potential of microbes for the recovery of radioactive nuclide. Table 3 depicts the comparative analysis about advantages of bioremediation with other remediate methods.
Table 3. Comparative Analysis of Bioremediation in Comparison to Conventional Remedial Methods
Advantages of Bioremediation as Compared to Conventional Remedial Methods |
Bioremediation takes advantage of the natural ability of microorganisms to extract chemicals from water, soil, and sediment using energy from sunlight. |
It is a cost-effective technique compared to other physicochemical treatment methods |
Less energy is required as compared to other technologies |
Often little to no residual treatment is required |
Bioremediation can be done on site and is often less expensive, and site disruption is minimal |
Soil stabilization and reduced water leaching and transport of organic compounds in the soil |
Typically, lower cost to implement |
Enhanced regulatory and public acceptance |
2 BIOREMEDIATION OF RADIONUCLIDE
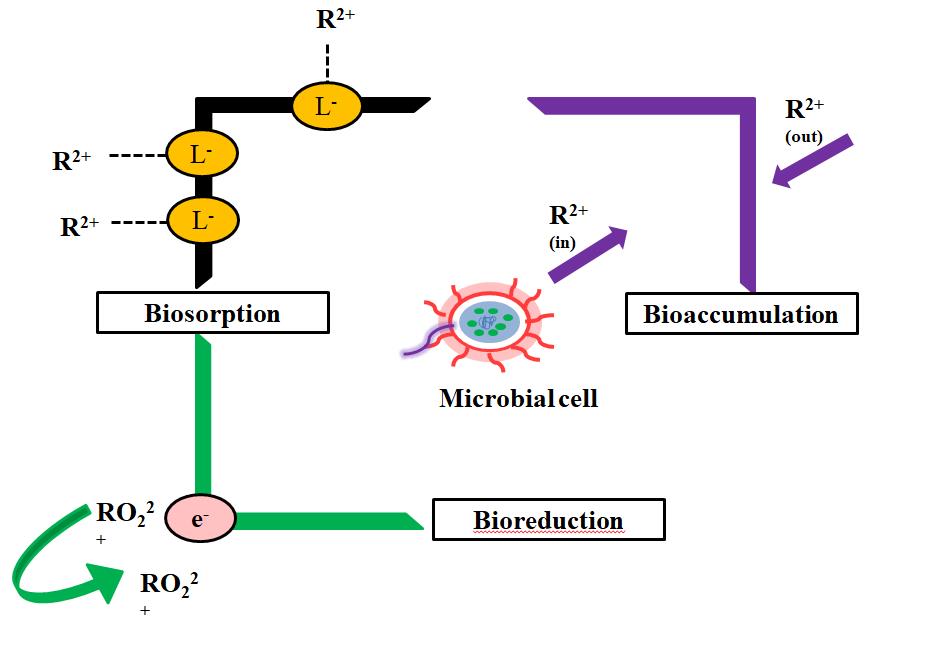
Microbes catalyze the conversion of inorganic and organic material naturally and can be utilized efficiently to solubilize or immobilize various forms of radioactive nuclides and toxic metals that are part of waste streams[10,21,22]. The basic mechanisms of microbial bioremediation of radionuclides are oxidation-reduction reactions, which affect the formal charges and their solubility, formation of complexes, bioaccumulation and biosorption, as depicts in Figure 1.
|
Figure 1. Mechanism of bioremediation of various radioactive nuclear wastes by biologically mediated reduction, sorption and accumulation in living system[22] (R2+ : Radioactive nuclide).
Microbial activities are greatly influenced by the capability of electron acceptance and donation[23-25]. Most microbes utilize oxygen as electron acceptor, while under anaerobic conditions nitrate, sulfate, metals, and carbon dioxide are widely used as exchange sources for electron capturers of microbes[26]. Under anaerobic conditions, the degree of precipitation and solubilization of radioactive nuclide is enhanced. Effective bioremediation of radioactive nuclides is predicated on the formation of complexes associated with physical, biological and chemical processes. The fundamental mechanisms include dissolution, oxidation, precipitation, reduction, sorption and leaching, and all these processes can reduce the toxicity and transfer the radioactive nuclides in biogeochemical cycle. All these basic mechanisms are realized via microbes, and bioprocess actively involved in the bioremediation of radioactive nuclides are designed to immobile them in consign to accelerate the recovery of radionuclides from waste stream[27,28]. Turick and Berry[29] studied microbial activity involved in the degradation of concrete at nuclear waste storage sites. Reena et al.[30] presented a complete database of bacteria and fungi, both wild kind and recombinant, which might be used in remediation of radioactive nuclide waste and has been created as ‘BioRadBase’. Xu and Zhou[31] noted that the great benefits of using microbes for the remediation of radioactive waste are high specificity, reusability more efficient, cost effective, minimum pollution and proper elimination of pollutant.
3 CLASSIFICATION OF BIOREMEDIATION
Bioremediation is further classified into three types (microbial, fungal and plant-based remediation) based on the type of biomass, as depicted in Table 1.
3.1 Microbial Remediation
The conversion of the radioactive nuclide into stable isotopes indirectly by the process of bacterial energy transfer is known as bacterial remediation. Radioactive nuclides are indirectly transformed by reducing and oxidizing agents produced by microbes, which can change their pH and their oxidation state. Bacterial remediation consists of different processes such as bioreduction, biosorption, bioaccumulations, and biomineralization involved in radionuclide transformation[16]. Bacteria can easily immobilize the radioactive nuclide by enzymes either directly or indirectly. The basic processes involved are bioreduction, biosorption, biomineralization and bioaccumulation.
3.1.1 Bioreduction
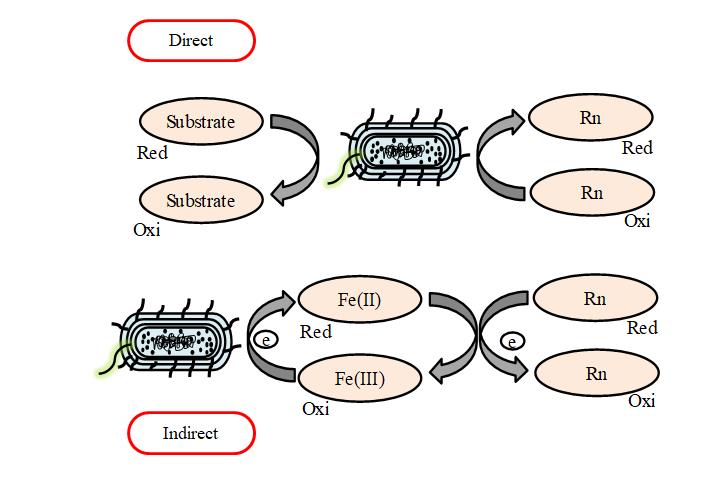
Bioreduction is a process in which bacterial species use redox potential and a reduction occur, through which soluble radionuclide becomes insoluble form. Bioreduction is a reliable technique because it is easy to operateunder mild environmental conditions and does not produce hazardous wastes[17]. Bioreduction is carried out by direct and indirect methods. In the direct enzymatic process, high oxidation state is converted to a less oxidation state by mean of anaerobic bacteria. In this process radionuclide are efficiently attached to binding sites on the surface of bacteria and they act as electron acceptor with ethyl lactate as the electron donor during anaerobic respiration. On the other hand, indirect bioreduction involves the indirect reduction of radionuclides to less soluble and less toxic species via sulfate-reducing bacteria. Some common example of both type bioreduction bacteria are Geobacter sulfurreducens, Shewanella putrefaciens, Desulfovibrio desulfuricans and Desulfovibrio vulgaris. The basic mechanism of bioreduction for both direct and indirect method is depicted in Figure 2.
|
Figure 2. Direct and Indirect enzymatic reduction of radionuclides by bacteria[22].
3.1.2 Biosorption
Biosorption, bioaccumulation and biomineralization are similar processes given their direct connection between the cell surface and radioactive nuclide[32]. All these are active processes dependent on energy transfer systems. Biosorption is a process in which positively charged radionuclide form complex with normally negatively chagred biomass either dead or alive[33]. Biosorption process involves the immobilization of the radioactive nuclide, which is species specific. Factors such as temperature, pH, aeration, growth phase of the cell, secretion of the exopolymer, and composition of the metabolites can affect biosorption. Moreover, the chemical interaction of extracellular biopolymers, electrostatic attraction and functional groups along with metal ions also affect biosorption[34]. Some bacterial species such as Citrobacter freudii and Firmicutes have significant capacities for biosorption. The carboxyl group in the cell wall of Citrobacter freudii is one of the most active sites of biosorption and has important effect on sorption[35]. They also revealed that dead cells display better sorption capability because the whole process was influence by functional groups other than the biological activity of the cell. The main disadvantage of biosorption is the rapid saturation of nuclide molecules and competitive desorption, which is caused by cations other than the targeted one competing for the cell's binding site[36]. As a result, biosorption is commonly used to remove low-concentration radionuclides from effluents[37].
3.1.3 Bioaccumulation
The process of bioaccumulation is defined as the uptake of radioactive nuclide into cell, where the cell complex is formed by positively charged radioactive nuclide and negatively charged cellular components in the form of small grains or precipitation[38]. The mechanism of bioaacumulation is based on formation of complex in which radioactive nuclide come into direct contact with some ligands such as phosphate, hydroxide, sulphate, forming insoluble substances inside the cell that can be easily removed from the solution. These insoluble forms are less harmful to the environment[39]. Micrococcus luteus is common example of bioaccumulation of strontium, forming complexes on the cell surface[17]. The process of bioaccumulation might be active or passive. Active bioaccumulation is a slow and more energy-depleting process that either depends or not on metabolism, while passive bioaccumulation is more promising due to limited nutrients. In contrast to active process it is a fast and less energy consuming process[40]. Both intracellular or extracellular accumulation is feasible. Intracellular accumulation of radionuclides is more prominent when existence or non-existence of respective radioactive nuclide influences the permeability of cell membrane[37].
In Citrobacter sp. and Halomonas sp., intracellular accumulation of U+6 was mostly in the form of phosphates such as hydroxophosphate, polyphosphates, or uranium hydrogen phosphate[41]. Amachi et al.[42] showed that the accumulation of iodide on the cell wall of soil Bacillus subtilis was shown to increase with the addition of glucose. Ozdemir et al.[43] studied bioaccumulation of U+6 by Bacillus vallismortis and found that after 72h of inoculation, bioaccumulate U+6 was at a concentration of 5mg/L. The amount of U (VI) accumulated was about 50mg/g (metal/dry bacteria), which was later analyzed by UV-Vis spectrophotometry. Another study on radionuclide exchange between uranium and a proton was conducted utilizing a separation factor, and it was shown that Gram-positive bacteria such as Micrococcus luteus, B. subtilis, and B. megatarium could accumulate Th in a solution containing both Thorium and Uranium[44]. Bioaccumulation is influenced by the molecular properties of the nuclide, as well as the potentials and characteristics of the bacteria present, as it includes rapid interactions with anionic groups in components of the cell surface[45]. It is also influenced by size and lipid content, with a reduction in size resulting in reduced surface area for accumulation[46]. The presence of capsules, slime, or S-layers has a significant impact on the bioaccumulation process. Polyphosphate bodies act on the nuclides after their accumulation. Metal tolerance and subsequent radiation tolerance have been linked to intracellular chelation processes[47]. However, according to Jiang et al.[48], the surface characteristics of bacterial cell walls determine their adsorption properties. Variations in cell wall composition between Gram-positive and Gram-negative bacteria had little effect on bioaccumulation[49].
3.1.4 Biomineralization/Bioprecipitation
Biomineralization is the formation of precipitate through the interactions of radioactive nuclide and microbial ligand[36], which is known as bio precipitation. The precipitation of radionuclides usually occurs in the form of carbonates or hydroxides. The oxidation state and valency of radionuclide have prominent effect on bio precipitation. The product from during biomineralization is a stable composite and biogenic material[50,51]. In microbial cell, the site of precipitation is known as the ‘nucleation site’ and it depends on the concentration of ligand produced by the cell. The precipitation of radionuclide is influenced by the ligands produced by microbes and biogenic formation of minerals[36]. Bacterial metabolism and secretions are prime factor that can change the pH and thus the pH in the vicinity of radionuclides[37]. Co-precipitation is another phenomenon where elements combine with metal oxide minerals during their precipitation[38]. Citrobacter and Serratiais are common example of biomineralization, which release phosphate ligand and form uranyl ions[52]. Table 4 summarized the mechanisms of radionuclide microbial bioremediation[53-65]. The use of these mechanisms of microorganisms for radioactive waste depends mainly on the individual capabilities of the species. Therefore, microorganisms appear again and again as they are the best choice for the remediation of radioactive wastes.
Table 4. Various Mechanisms of Radionuclide Microbial Bioremediation[53]
Mechanism |
Radionuclide |
Microbes |
Ref. |
Metabolism-dependent; Sulphide precipitation Transport Intracellular compartmentation Sequestration by proteins, peptides Immobilization by chemical reduction Siderophore complexation |
U+6, Ga+3, Cr+3, Pu+4, Sc+, In, Ni, U Th |
Shewanella putrefaciens Desulfovibrio desulfuricans Desulfotomaculum sp. Geobacter metallireducens Aspegillus niger |
[54] |
Enzymatic; Direct Indirect
|
U+6 |
G. sulfurreducens, Clostridium sp., S. putrefaciencs, Desulfovibrio vulgaris, D. Desulfuricans |
[37,50,51,54-58] |
Biomineralization
|
Cr+6, As sulphides, phosphates |
Citrobacter, Acinetobacter johnsonii, Klebsiella planticola, Pseudomonas aeruginosa, P. putida, Salmonella enterica, Escherichia coli |
[50-51] |
Biostimulation |
Pu+6, U+6, Np+5, Tc+7 |
Geobacter, Arthrobacter ilicis |
|
Bioaugmentation Dissimilatory reduction |
Hg+2, As+5, Fe+3, U+6, Co+3, Cr+6 Tc+7 |
Streptomyces, Aciditiobacillus ferrooxidans, P. putida, Ralstonia eutropha, Pyrobaculum islandicum, Geobacter, Sulfospirillum barnesii, Rhodanobacter sp., Desulfuromusa ferrireducens, Deinococcus radiodurans, Sulfurospirillum barnesii |
|
Biotransformation Biosorption Bioaccumulation |
Pb+2, Zn+2, Cd+2, Sr+2, Th, U+6 |
Pseudomonas mendocina, Bacillus subtilis, Arthrobacter spp., Rhizopus arrhizus, A. nicotianae, Micrococcus luteus, B. vallismortis |
[39,43,50,59-62] |
Genetically modified microorganisms |
Uranyl nitrate, Co, Hg+2 |
D. radiodurans R1, Escherichia coli, R. eutropha, |
[42,51,55,63-65] |
Bioprecipitation |
Uranyl phosphate |
D. radiodurans |
[60] |
3.2 Plant Remediation
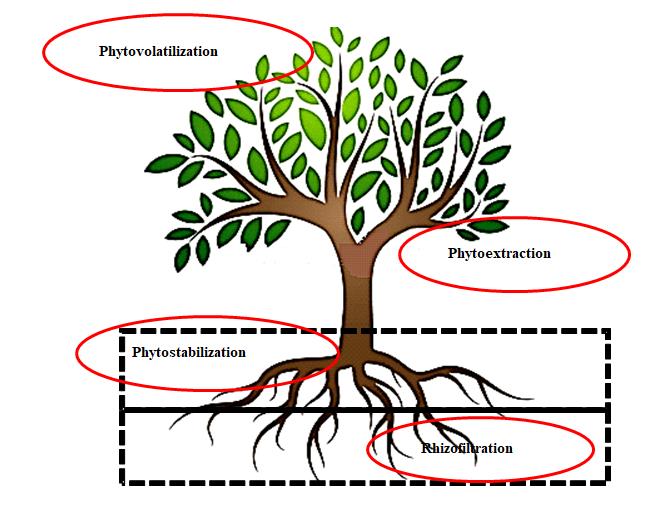
The term "phytoremediation" refers to a group of remediation techniques that employ plants to clean or partially clean contaminated sites or reduce the danger of toxins. Phytoremediation is also known as green remediation, botano-remediation, agro-remediation, and vegetative remediation[66,67]. Plants have the capability to uptake pollutants in the environment through the root system that provides a larger surface area, ease mobilization, and detoxification of contaminants within plants by using a variety of mechanisms such as elimination, containment, and degradation. Such plant characteristics have been employed to effectively remove radioactive waste. The microbial population associated with the plant and their interactions play a crucial role in maintaining the health of the plant. These interactions inhibit phytopathogens by releasing compounds that promote growth, increase the availability of nutrients, encourage detoxification (e.g., degradation, sequestration, and volatilization of pollutants), and enhance stress tolerance by introducing systematically acquired host resistance. Leaves, stems, and roots are habitats for a wide range of microbes that easily degrade toxic pollutants, which improves the treatment process. As illustrated in Figure 3, plants use various processes to absorb organic and inorganic pollutants, including phytoextraction, phytostabilization, phytodegradation, phytovolatilization, and rhizofiltration. These mechanisms constitute the basis of phytoremediation technology.
|
Figure 3. Various process involves in plant remediation of radionuclides[52].
3.2.1 Phytoextraction
Phytoextraction is a process in which radioactive nuclide concentrates in the shoots of the plants. The radionuclides are transferred from the roots to the shoots through the vascular bundles, forming a complex with the biomass of the shoots, converting the radionuclides to a less toxic form[66,67]. The benefit of this process is the easy removal of radionuclides without disturbing the soil structure and its fertility. In the chernobyl exclusion zone, the most abundant radioactive nuclide is frequently concentrate by common heather or amaranths species.
3.2.2 Rhizofiltation
In rhizofiltration, radionuclides are adsorbed and precipitated in the roots of plants, but their effectiveness depends entirely on the pH[66,68]. Cseium-137 and Stroncium-90 both are significantly adsorbed in the roots of some aquatic plants and algae species such as Cladophora and Elodea. At present, a large number of ponds are synthetically designed for this process, in which different aquatic plants and algae are grown to remove radionuclides[69]. Sunflower is the most efficient plant used for rhizofiltration because it can adsorb up to 95% radioactive nuclide from waste stream within two days.
3.2.3 Phytovolatilization
Phytovolatilization is a process involving the volatilization of radioactive nuclide in the form of less toxic substance[66,70]. Phytovolatilization process proceeds with transpiration of radioactive nuclide into atmosphere. Radionuclides are not removed during this process, but the process effectively releases radionuclides in the form of volatile substances that are less toxic to the environment. The process is cost-effective compared to other bioremediation processes, and its main advantage lies in its non-disruption of soil structure and fertility.
3.2.4 Phytostabilization
During phytostabilization, the roots of plants are involved in the immobilization of radioactive nuclide[66]. The basic mechanism of the plant stabilization process is the adsorption and precipitation of radionuclides in the roots of plants[71]. The main benefit of this process is that radioactive nuclides are immobilized in roots and can be easily removed if the waste stream of radioactive nuclide is not affected by leaching and soil erosion. Nevertheless, the excessive use of fertilizer in the soil for the restoration of effected area may compromise the benefits of this process[67]. Table 5 summarized the mechanism of phytoremediation of radionuclide[67-99]. Recently, the development of phytoremediation technology has received extensive attention and it is expected that it will eventually occupy an important place in environmental remediation. It is envisaged that phytoremediation of radioactive waste will be an essential component of its environmental management and radioactive waste risk reduction.
Table 5. Various Mechanisms of Radionuclide Phytoremediation[66]
Mechanism |
Radionuclide |
Plant species |
Ref. |
Phytoextraction |
99Tc |
Deciduous forest, Triticum, Lolium, Mixed grass species |
[72-74] |
Phytoextraction |
90Sr |
Pinus Radiata pine, ponderosa pine, Vitis, Agrostis, Sorghum, Panicum, Paspalum Mycorrhizal association, Salix (willow), Calotropis, Vetiveria, Chromolaena, Cannabis |
[75-83] |
Rhizofiltration |
90Sr |
Catharanthus, Helianthus, Brassica, Eichhornia, |
[84-87] |
Phytoextraction |
90Co |
Melilotus, Sorghum, Trifolium |
[88] |
Phytoextraction |
222Rn |
Helianthus, Festuca, Zea |
[89] |
Phytoextraction |
226Ra |
Helianthus, Festuca, Zea |
[89] |
Phytoextraction |
238,235U |
Helianthus, Brassica, Phaseolus, Pisum, Beta, Brassica, Amaranthus |
[84,90,91] |
Rhizofiltration |
238,235U |
Brassica, Chenopodium, Eichhornia |
[84,92] |
Rhizofiltration |
239,240Pu |
Sargassum, Hordeum, Soya |
[93,94] |
Rhizofiltration |
237Np |
Triticum, Lolium |
[95] |
Phytoextraction |
210Po |
Tabaccum (tobacco) |
[96] |
Phytoextraction |
137Cs |
Brassica, Spinacia, Lactuca, Raphanus Salix (willow), Calotropis, Vetiveria Vetiveria , Soya, Pisum, Avena Mycorrhizal association |
[78,79,81,97,98] |
Rhizofiltration |
137Cs |
Helianthus, Eichhornia, Catharanthus |
[84-86,99] |
3.3 Fungal Remediation
Fungal bioremediation, called myco-remediation, is considered more effective for radioactive nuclide removal from waste streams when compared with bacteria fungi[100,101]. The basic mechanism of myco-remediation is identical to biosorption. The cell wall of fungi is negatively charged, and positive radioactive nuclide form complex on its surface[102,103]. Some common species of fungi that involves in biosorption are Xerocomus, Cladosporium Paecilomyces, and Penicillium, which effectively adsorb radioactive nuclide such as Promethium-239 and Amercium-241 because these fungal species use the radioactive energy of these nuclide for their growth[104]. Table 6 summarized radionuclides fungal remediation[105-110]. The advantage of using fungi for remediation is that they are natural decomposers and secrete enzymes that dissolve toxic contaminants without any hazardous effects. This approach allows the development of an eco-friendly way to treat and extract/recover precious radionuclides.
Table 6. Various Mechanisms of Radionuclide Myco-remediation[105]
Mechanism |
Radionuclides |
Fungi |
Ref. |
Biosorption |
233U |
Rhizopus arrhizus, Gibberella fujikuroi, G. fujikuroi NCIM 665, G. saubinetti NCIM 851, richoderma harzianum, Rhodotorula glutinis |
[106,107] |
Biosorption |
239Pu |
Rhizopus arrhizus, Gibberella fujikuroi, G. fujikuroi NCIM 665, and G. saubinetti NCIM 851 |
[106] |
Biosorption |
241Am |
Rhizopus arrhizus |
[108] |
Biosorption |
144Ce |
Rhizopus arrhizus, Gibberella fujikuroi, G. fujikuroi NCIM 665, and G. saubinetti NCIM 851 |
[106] |
Biosorption |
147Pm |
Rhizopus arrhizus, Gibberella fujikuroi, G. fujikuroi NCIM 665, and G. saubinetti NCIM 851 |
[106] |
Biosorption |
152,154Eu |
Rhizopus arrhizus, Gibberella fujikuroi, G. fujikuroi NCIM 665, and G. saubinetti NCIM 851 |
[106] |
Bioaccumulation |
152,154Eu |
Saccharomyces cerevisiae |
[109] |
Biosorption |
137Cs |
Ladosporium, cladosporoides |
[110] |
Biosorption |
90Sr |
Ladosporium, cladosporoides |
[110] |
4 BIOREMEDIATION OF ACTINIDES AND THEIR FISSION PRODUCTS
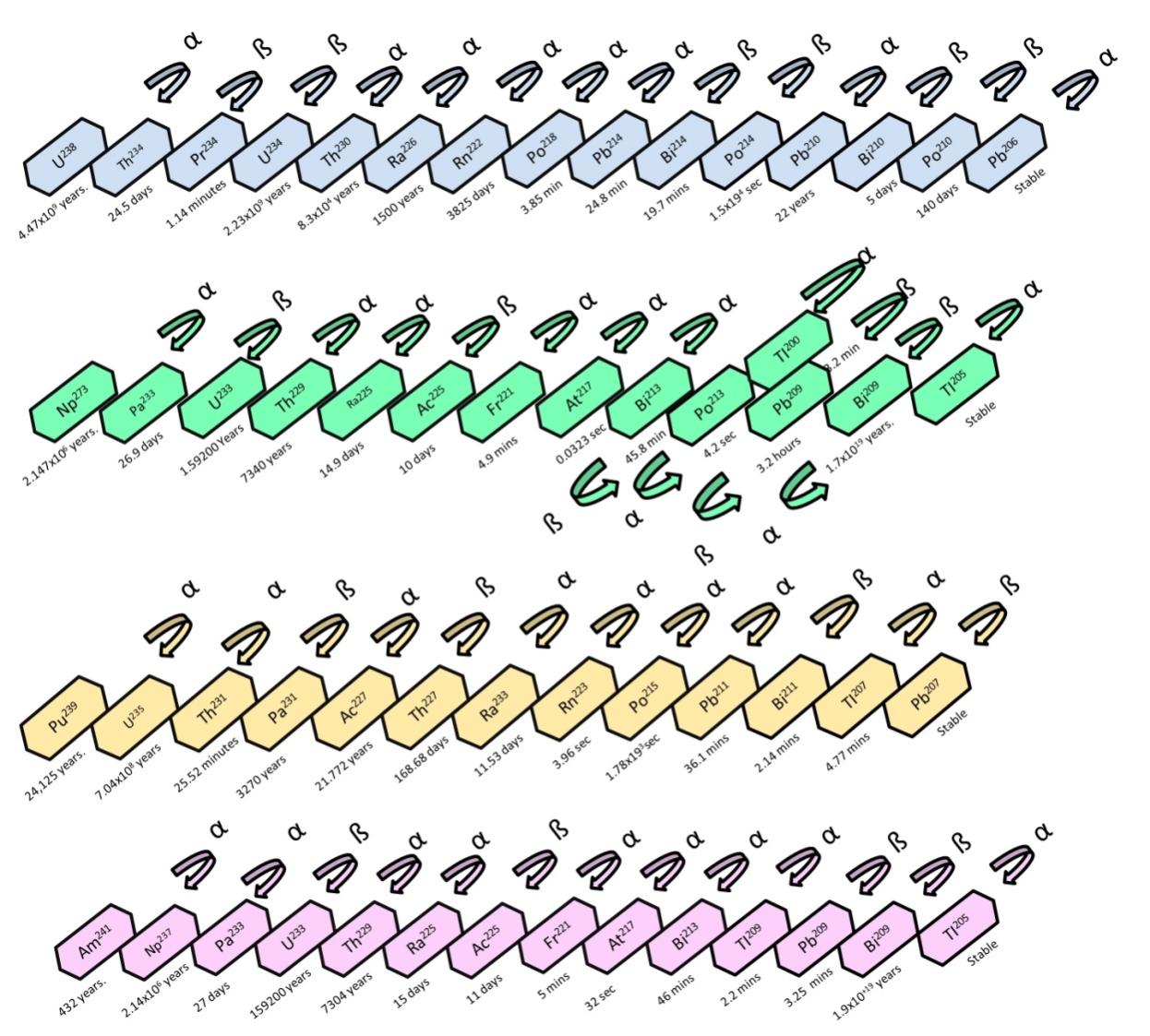
The second series of f-block elements with valance shell electronic configuration [Rn] 5f1-14, 6d0-10, 7s2 are known as Actinides[111]. All elements in the actinide series are radioactive in nature, and radioactive decay releases a significant amount of energy. The radioactive decay of various actinides is depicted in Figure 4. The most abundant and naturally occurring actinides on Earth are uranium and thorium, while plutonium and others are artificially synthesized. Exposure to these radionuclides is a concern for health and ecology. In biotechnological applications, microbes possess a massive capability for undergoing sustainable manipulations and are, therefore, used for excretion of radionuclides from environment. Radionuclides are non-destroyable but transformable[50]. Bioremediation of various actinides is discussed as follows.
|
Figure 4. Radioactive decay series of various actinides Uranium238, Neptunium273, Americam241 and Plutonium253[111].
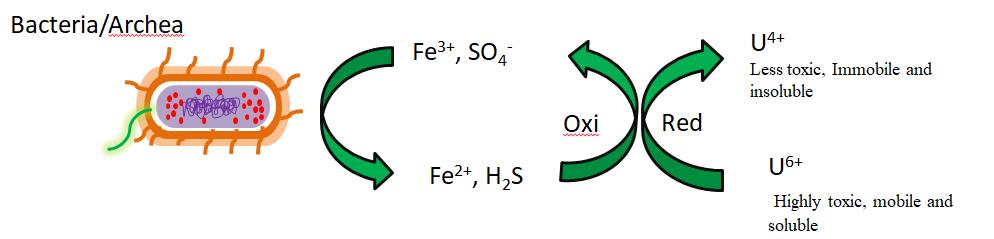
In nature, about 99% of uranium exists as Uranium-238 with a half time of 4.47×109 years[111,112]. Uranium-238 is an alpha particles emitter radioactive isotope abundantly found in nuclear wastes including nuclear weapon generation and recycling as well as during nuclear fuel production. There are several microorganism that shows strong affinity to remove uranium from aqueous solutions by using some enzymes involving a reduction process that converts Uranium +6 soluble species into Uranium +4 insoluble species via precipitation of Uranium +4 with the use of some phosphate ligands or by forming complex with the surface of cell, known as biosorption[113,114]. The most common microbes, including bacteria, are Geobacter metallireducens ferric reducing bacteria, Shewanella oneidensis[115,116], Clostridiu[21], Desulfovibrio desulfuricans and Desulfovibrio vulgaris that show affinity with uranium and can remove it through complexolysis, redoxolysis, bioprecipitation and biosorption. There are two classes of microorganisms that have affinity with uranium. The first class consists of bacteria that conserve energy during reduction of Uranium +6 to Uranium +4 for their anaerobic UO22+ growth, and the second class consists of bacteria without the ability to store energy for their metabolism during reduction. Geobacter metallireducen and Shewanella oneidensis are most common example of bacteria that conserve energy for their growth during reduction process[117]. On the other hand, Clostridium, Desulfovibrio desulfuricans and Desulfovibrio vulgaris are common example of bacteria that cannot store energy for their metabolism during reduction of radioactive nuclides[114,118,119]. The basic mechanism of all bacteria is the presence of several types of cytochromes on the cell surface, which are involved in the reduction of uranium and act as electron donors[120,121]. Figure 5 illustrates well the reduction of Uranium +6 to Uranium +4 through interaction with bacteria.
|
Figure 5. Microbial reduction of uranium[52].
Neptunium is the fourth element in the actinide series after uranium. Neptunium is highly radioactive nuclide emitting alpha radiations with a long half-life about 214×106 years, and human exposure to it will cause serious health issues[122,123]. Currently, neptunium is majorly produced due to anthropogenic activities including nuclear fuel explosion and nuclear weapons. The most common oxidation state of neptunium is Neptunium +4 and Neptunium +5. Neptunium +4 is an insoluble species that readily precipitates as hydroxide and carbonates, while neptunium +5 is soluble species as it combines with oxygen and form a soluble species, neptunium oxide, that is abundantly found in aquatic media[124-126]. Several classes of microbes have affinity with neptunium including marine algae and bacteria. Pseudomonas aeruginosa, Streptomycensvirido chromogenes, Scenedesmus obliquus and Micrococcus are some common marine alage that involve the removal of neptunium by forming complex on their cell surface. Pseudomonas fluorescens is well-known marine algae that efficiently adsorb neptunium +5 from aqueous media at neutral pH by forming complexes on its surface[127]. Similarly, some bacteria are also known for the removal of soluble oxide species from aqueous media to insoluble species neptunium +4 easily by using enzymes and complexing activities. Shewenella putrefaciens, Citrobacter species, Desulfovibrio desulfurains are some common bacteria that reduce neptunium +5 to neptunium +4 and they easily precipitate out as neptunium phosphate in the presence of phosphates.
Plutonium is another trans-uranium artificial element. Plutonium-238 is considered a highly radioactive isotope emitting beta radiations and has a half-lifeof 87.7 years. Plutonium is considered most debatable actinide element due to its dual uses in disrupting nuclear weapon and nuclear energy generation, which accounts for its large presence in the nuclear waste stream. The most common oxidation state of plutonium is +4, and plutonium +3, +5 and +6 are also stable species. Radioactive decay series of Plutonium-238 is described in Figure 4. Limited data is available on microbial interaction with plutonium. Microbial interaction with plutonium and its reduction from plutonium +4 to plutonium +3 has been reported by using ferric reducing bacteria, but re-oxidation of plutonium +3 occurs spontaneously. The reduction of plutonium +5 and plutonium +6 to plutonium +4 via enzymes with the aid of bacteria has also been done by using Shewenella putrefaciens, Shewenella oneidensis[128]. These bacteria can convert the soluble species of plutonium into insoluble species by mean of reduction and then easily precipitate out with some inorganic complexion ligands; however, it has been reported that oxidation state of plutonium is eventually altered by microbial system and that the solubility of plutonium depends on the redox potential and pH of the solution. A case study investigated the reduction of plutonium from plutonium +4 to plutonium +3 by using microorganisms, in which all electron acceptors and markers were observed, and it revealed that the release of low level of plutonium from deposits with ferric reduction, indicating that plutonium is resistance to reduction mobilization[129,130]. Americium is also a trans-uranium element. Americium-241 emits alpha particles and decays into Neptunium-237 with a half-life of 432.2 years. Americium has several oxidation states from +2 to +7 but Americium +2 is usually found in solid state. Several microbes play an important role in interacting with americium, reducing Americium +3 to Americium +2, and adsorbing efficiently on their surface. Some common microbes involved in the reduction from the recovery of americium from devastate include Escherichia coli, Candida utilis, Ochrobactrumanthropi, Flavobacterium, Pseudomonas gladioli and Chryseobacteriumindologenes[131], Rhizopusarrhizus[106]. The radioactive decay of all these neptunium, plutonium and americium are described in Figure 4.
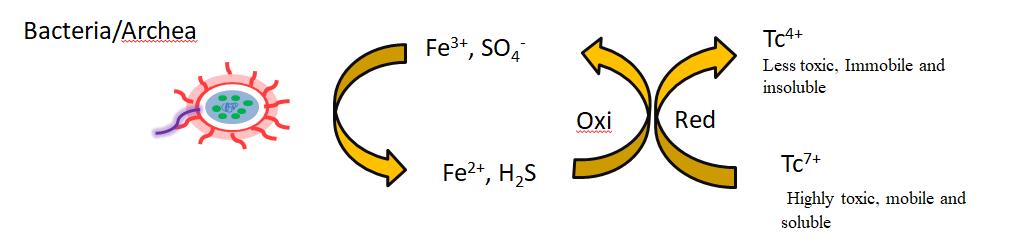
Technetium-99 is beta emitter radioactive nuclide with a half-life of 2.13×105 years. Technetium-99 is important component of nuclear waste and is highly radioactive[132]. The chemistry of technetium is totally depending upon environmental solubility. The most common oxidation state of is technetium +7, which exists as technetium tetra oxide (TcO4) and is highly mobile species in environment because it has little adsorption on the cell surface due to its high solubility. However, through the use of some microbes, technetium +7 is converted to technetium +4 by the reduction process and forms insoluble complex technetium dioxide (TcO2)[133-135]. Most common examples of microbes that involve in bioreduction of technetium includes Shewanella putrefaciens, Geobacter metallireducens, Rhodbacter sphaeroides, Pseudomonas denitrificans, Pseudomonas species and Escherichia coli[136,137]. Escherichia coli is considered the most suitable for the bioreduction of technetium. Firstly, in anaerobic culture, Escherichia coli reduces the technetium +7 to technetium +4 and precipitates it on the surface of the cell[138]. The use of hydrogenase allows them to form formate hydrogenase complexes[139]. Furthermore, Desulfovibrio desulfuricans and other related strains also use formate as an electron donor to reduce technetium and form insoluble complexes on the surface[140]. The reduction of technetium +7 to technetium +4 by interaction with bacteria is well illustrated in Figure 6.
|
Figure 6. Microbial reduction of Technetium[52].
Cesium is a radioactive nuclide produced as a fission product. Cesium-137 has a half-life of about 30 years. In the environment, large amounts of cesium are released as a result of nuclear explosions. In the environment, the most common form of cesium is cesium-137 in the Fukushima, Chernobyl and Goiania accidents. Microbes play a key role in the bioaccumulation of cesium, but pre culture studies revealed insufficient uptake of cesium by microorganisms. However, the more efficient uptake of cesium by microorganisms is highly promising[141]. This is highly similar to the behavior of potassium ions uptake given the similar metabolic transport system of both cations. Most studies on the uptake of cesium ions are shallow. From aqueous media, microorganisms simply adsorb them on their surface by forming complex at alkali pH. Strontium-90 is a fission product that undergoes beta particle emission with a half-life of 29 years. This fission product is formed during exploitation of nuclear reactors. It is frequently emitted as cesium-137 because it is less volatile but is considered the most dangerous radionuclide pollutant. Therefore, its accumulation is necessary because it present in significant amount in nuclear explosion waste stream. Microbes perform a vital role in the removal of radionuclides as they efficiently adsorb strontium ions on their surface by forming complexes. Microccous leteusis is a common example for strontium adsorption. Strontium binding site is present on the surface of Microccous leteus, and Strontium ions easily displace by divalent ion and chelating agents[142,143]. Radium has the highly stable isotope Radium-226 with a half-life of 1600 years. Radium is usually associated with uranium ore and also dissolved with uranium at the uranium milling site, so it can be precipitated by using barium sulfate and barium chloride to form a radioactive barium sulfate/radium sludge. Its supernatant is released into the environment and contains some radionuclide of radium. Microbes are important to remove radium-226 by co-precipitation with other metallic ions through reduction. Actinobacteria throbacter, is a well-known species that remove mobile manganese and Radium +2 by co-precipitation, while bacteria reduce Manganese +4 to Manganese +2[58,142].
5 EXISTING RADIOACTIVE NUCLIDE WASTE MANAGEMENT PROGRAMS AND FUTURE STRATEGIES IN DEVELOPING COUNTRIES
A program of action or plan designed by a government or different organizations to influence decisions is called a policy. At the beginning of the nuclear era, countries that started introducing nuclear energy were ignorant about nuclear waste and its remediation. To deal with destruction of radionuclides, most of the countries are developed and urbanized and they have adopted basic strategies and methods for the disposal of nuclear waste. Some countries disposed the radioactive nuclide on site storage without proper management for their disposal at national level. In 1995, International Atomic Energy Agency, published the basic principle for proper disposal of radionuclide waste and its ethical issues to reduce the hazardous effects of radioactive nuclide on human health and the environment.
According to this statement, radioactive nuclide waste should be properly managed to minimize the risk to human health and provide an appropriate level of environmental protection. This management practice also ensures that radioactive nuclide waste had little effect on the next generation. Radioactive nuclide waste should be handled within proper and legal national framework, including independency of regulatory function and clear allocation of responsibilities. All the major steps in production and the management policy radionuclide waste are interdependent, and the safety facilities of the radionuclide waste management plan will be properly established during their life cycle[144].
The fundamental ideology associated with radionuclide waste disposal can be imposed on all types of radionuclide, despite their different chemical and physical characteristics and origins. Based on these principles, all countries have their rules and regulations as well as their national policies that describe the basic requirements and aims for the legislative and regulatory bodies that includes operative measures and administration[145]. These fundamental principles explain the situation, state priority, structural, financial, and human resources. In 1922, International atomic energy agency recognized the basic principles and essentials of the advanced policies related to radionuclide waste management. To obtain better policy principles, national waste management plans should be adapted to their implementation in practice and to changing conditions in the country or the world.
The rules for radionuclide waste management, once developed, need to be implemented in practice. Many steps are involved in the implementation. The first is the development of a management strategy, the assignment of strategic responsibilities, and then the acquisition of availability that will help expand the policy. The International Atomic Energy Agency has developed an availability checklist that aids in the development of appropriate strategies for other countries. This checklist includes an assessment of the waste management system, the classification of radionuclides, and an assessment of the sources of radionuclide waste. The second step of the radionuclide waste implementation policy is to identify the endpoints of the waste. Finally, optimal management strategy is developed and responsibilities are assigned for their implementation. There are two options for strategy development: One-stage method and the two-stage methods. The one-stage method is also called nationalized plan that has single waste operator to evaluate the strategy and its implementation. The two-stage method starts with the description of general management policy provisions by an organization such as the government, while in a second step these management policies are implanted by some private organization such as a single company waste operator.
To dispose of all radioactive nuclide waste in developing countries in a safe and sustainable way, National Energy Agency provides considerable assistance. To this end, the National Energy agency has issued strategic management plans, which is the responsibility of the Radionuclide Waste Management Committee (RWMC), to understand the basic problems and challenges faced by all developing countries during the disposal of radioactive nuclides. The recognized designed area of interest integrated the following.
● Development of a proper and sustainable management system that includes financing;
● Disposal of radioactive waste along spent fuel transportation through optimized and robust roadmaps development;
● License must be approved for geological repositories for high and low-level waste;
● Execution of deep geological disposal for industries;
● Decommissioning is effective;
● Long term preservations record and knowledge management and memory;
● Effective executive of all types of radionuclide waste despite of its origin.
6 CONCLUSION
With the latest advances in atomic energy area, radionuclides are generated to meet the demand for nuclear energy and the safe disposal of these radionuclide wastes is a current issue. The release of radioactive nuclide waste from nuclear sites and its spent sources including mining activities, industrial activities, land fillings, agrochemical waste, nuclear weapons recycling, nuclear weapon and nuclear energy generation causes deteriorate effect on environment as well as on human health. These radioactive nuclides majorly consist of high-energy beta and gamma radiation emitters that cause serious health issues such as cancer after excessive exposure to them. Natural resources such as water and soil are also at risk of contamination by radionuclide leaching due to the geological map of mining activities. Hence, to deal with these serious issues, there exists an urgent need to develop nuclear waste management programs that provide an effective route for the disposal of radioactive nuclear waste. Historically, different methods such as solvent extraction, precipitation, ion exchange, and electrochemical process for the effective removal of radio nuclide from waste nuclear stream have been adopted. However, in the recent era, bioremediation demonstrates a great potential to use microorganisms/biomass for the removal and treatment of radioactive nuclide from waste streams, as they are easy to cultivate, economical, and non-chemical pollutants. Therefore, biotechnology plays an important role in different industrial sectors and for environmental protection. In this review, different types of biomass such as bacteria, fungi and plants were reported for the effective treatment and the restoration of radioactive nuclides. All three modes of bioremediation serve as a prime candidate for the management of radioactive waste. Moreover, to dispose of all radioactive nuclide waste in developing countries in a safe and sustainable way, an efficient strategy for radioactive nuclide waste management was also necessitated.
Acknowledgements
Not applicable.
Conflicts of Interest
The author declared no conflict of interest.
Author Contribution
Cheema HA studied, wrote, and reviewed this article.
References
[2] Khan NT. Radioactivity: An introduction to mysterious science. J Phys Chem Bio Phys, 2017; 7: 2161-2398. DOI: 10.4172/2161-0398.1000254
[3] Adebiyi FM, Ore OT, Adeola A et al. Occurrence and remediation of naturally occurring radioactive materials in Nigeria: A review. Environ Chem Lett, 2021; 19: 3243-3262. DOI: 10.1007/s10311-021-01237-4
[4] US EPA, In situ treatment technologies for contaminated soil, EPA 542/F-06/013. US EPA: Washington, USA, 2006.
[5] Jasaitis D, Klima V, Pečiulienė M et al. Comparative assessment of radiation background due to natural and artifcial radionuclides in soil in specific areas on the territories of state of Washington (USA) and Lithuania. Water Air Soil Pollut, 2020; 231: 1-10. DOI: 10.1007/s11270-020-04730-8
[6] Cazzola P, Cena A, Ghignone S et al. Experimental system to displace radioisotopes from upper to deeper soil layers: Chemical research. Environ Heal, 2004; 3: 1-10. DOI: 10.1186/1476-069X-3-5
[7] IAEA. Environmental contamination from uranium production facilities and their remediation. Proceedings of an International Workshop on Environmental Contamination from Uranium Production Facilities and Their Remediation, Vienna, Austria, 11-13 February 2004.
[9] Baca TE, Florkowski T. The Environmental challenges of nuclear disarmament. Springer: Heidelberg Berlin, New York, 2000.
[10] Lloyd JR, Renshaw JC. Microbial transformations of radionumlides: fundamental mechanisms and biogeochemical implications. Met Ions Biol Syst, 2005; 44: 253-260. DOI: 10.1201/9780849346071
[11] Napier BA, Reed PR. Alternative conceptual models for assessing food chain pathways in biosphere models. U.S. Nuclear Regulatory Commission Office of Nuclear Regulatory Research: Washington, USA, 2006.
[12] The Royal Society. The health hazards of depleted uranium munitions. The Royal Society: London, UK, 2001.
[13] El-Gamal H, Hussien MT, Saleh EE. Evaluation of natural radioactivity levels in soil and various food stufs from Delta Abyan, Yemen. J Radiat Res, 2019; 12: 226-233. DOI: 10.1080/16878507.2019.1646523
[14] Duggal V, Rani A, Mehra R et al. Assessment of natural radioactivity levels and associated dose rates in soil samples from Northern Rajasthan, India. Radiat Prot Dosim, 2014; 158: 235-240. DOI: 10.1093/rpd/nct199
[15] Chung AP, Lopes A, Nobre MF et al. Hymenobacterperfusus sp. nov., Hymenobacterflocculans sp. nov. and Hymenobactermetalli sp. nov. three new species isolated from an uranium mine waste water treatment system. Syst Appl Microb, 2010; 33: 436-443. DOI: 10.1016/j.syapm.2010.09.002
[16] Akob DM, Mills HJ, Kostka JE. Metabolically active microbial communities in uranium-contaminated subsurface sediments. FEMS Microb Ecol, 2007; 59: 95-107. DOI: 10.1111/j.1574-6941.2006.00203.x
[17] Phillips RW, Wiegel J, Berry C J et al. Kineococcusradiotolerans sp. nov., a radiation-resistant, gram-positive bacterium. Int J Syst Evol Microb, 2002; 52: 933-938. DOI: 10.1099/00207713-52-3-933
[18] Dozol M, Hagemann R. Radionuclide migration in groundwaters: Review of the behaviour of actinides. Pure Appl Chem, 1993; 65: 1081-1102. DOI: 10.1351/pac199365051081
[19] Favre-Réguillon A, Dunjic B, Lemaire M et al. Synthesis and evaluation of resorcinol-based ion-exchange resins for the selective removal of cesium. Solvent Extr Ion Exch, 2001; 19: 181-191. DOI: 10.1081/SEI-100001382
[20] Atlas RM. Bioremediation. Chem Eng New, 1995; 73: 32-42. DOI: 10.1128/9781555817596
[21] Francis AJ. Microbial transformations of radioactive wastes and environmental restoration through bioremediation. J Alloy Comp, 1994; 213: 226-231. DOI: 10.1016/0925-8388(94)90908-3
[22] Ilyas S, Lee J. Bioleaching of metals from electronic scrap in a stirred tank reactor. Hydrometallurgy, 2014; 149: 50-62. DOI: 10.1016/j.hydromet.2014.07.004
[23] You J, Solongo SK, Gomez-Flores A et al. Intensified bioleaching of chalcopyrite concentrate using adapted mesophilic culture in continuous stirred tank reactors. Bioresour. Technol, 2020; 307: 123181. DOI: 10.1016/j.biortech.2020.123181
[24] Ilyas S, Bhatti HN, Bhatti IA et al. Bioleaching of metal ions from low grade sulphide ore: Process optimization by using orthogonal experimental array design. Afr J Biotechnol, 2010; 9: 2801-2810.
[25] Iyas S, Ru'an CHI, Lee JC et al. One step bioleaching of sulphide ore with low concentration of arsenic by Aspergillus niger and Taguchi orthogonal array optimization. Chin J Chem Eng, 2012; 20: 923-929. DOI: 10.1016/S1004-9541(12)60419-4
[26] Naseem Z, Bhatti HN, Sadaf S et al. Sorption of uranium (VI) by Trapa bispinosa from aqueous solution: Effect of pretreatments and modeling studies. Desalin Water Treat, 2016; 57: 11121-11132. DOI: 10.1080/19443994.2015.1041059
[27] Elias DA, Krumholz LR, Wong D et al. Characterization of microbial activities and U reduction in a shallow aquifer contaminated by uranium mill tailings. Microb Ecol, 2003; 46: 83-91. DOI: 10.1007/s00248-002-1060-x
[28] North NN, Dollhopf SL, Petrie L et al. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl Environ Microb, 2004; 70: 4911-4920. DOI: 10.1128/AEM.70.8.4911-4920.2004
[29] Turick CE, Berry CJ. Review of concrete biodeterioration in relation to nuclear waste. J Environ Radioact, 2016; 151: 12-21. DOI: 10.1016/j.jenvrad.2015.09.005
[30] Reena R, Majhi MC, Arya AK et al. BioRadBase: A database for bioremediation of radioactive waste. Afr J Biotechnol, 2012; 11: 8718-8721. DOI: 10.5897/AJB12.020
[31] Xu Y, Zhou NY. Microbial remediation of aromatics-contaminated soil. Front Environ Sci Eng, 2017; 11: 1-9. DOI: 10.1007/s11783-017-0894-x
[32] Van Hullebusch ED, Lens PNL, Tabak HH. Developments in bioremediation of soils and sediments polluted with metals and radionuclides. 3. Influence of chemical speciation and bioavailability on contaminants immobilization/mobilization bio-processes. Rev Environ Sci Bio, 2005; 4: 185-212. DOI: 10.1007/s11157-005-2948-y
[33] Prakash D, Gabani P, Chandel AK et al. Bioremediation: a genuine technology to remediate radionuclides from the environment. Microb Biotechnol, 2013; 6: 349-360. DOI: 10.1111/1751-7915.12059
[34] Dobrowolski R, Szcześ A. Czemierska M et al. Studies of cadmium (II), lead (II), nickel (II), cobalt (II) and chromium (VI) sorption on extracellular polymeric substances produced by Rhodococcus opacus and Rhodococcus rhodochrous. Biores technol, 2017; 225: 113-120. DOI: 10.1016/j.biortech.2016.11.040
[35] Xie S, Yang J, Chen C et al. Study on biosorption kinetics and thermodynamics of uranium by Citrobacter freudii. J Environ Radioact, 2008; 99: 126-133. DOI: 10.1016/j.jenvrad.2007.07.003
[36] Jabbar T, Wallner G. Biotransformation of radionuclides: Trends and challenges. Radionuclides Environ, 2015; 169-184. DOI: 10.1007/978-3-319-22171-7_10
[37] Selenska-Pobell S. Diversity and activity of bacteria in uranium waste piles. Radioactivity Environ, 2002; 2: 225-254. DOI: 10.1016/S1569-4860(02)80037-7
[38] Tabak HH, Lens P, van Hullebusch ED et al. Developments in bioremediation of soils and sediments polluted with metals and radionuclidese. Rev Environ Sci Biotechnol, 2005; 4: 115-156. DOI: 10.1007/s11157-005-2169-4
[39] Haferburg G, Kothe E. Microbes and metals: interactions in the environment. J Basic Microb, 2007; 47: 453-467. DOI: 10.1002/jobm.200700275
[40] Velásquez L, Dussan J. Biosorption and bioaccumulation of heavy metals on dead and living biomass of Bacillus sphaericus. J Hazard Mat, 2009; 167: 713-716. DOI: 10.1016/j.jhazmat.2009.01.044
[41] Francis AJ, Gillow JB, Dodge CJ et al. Uranium association with halophilic and non-halophilic bacteria and archaea. Radiochim Acta, 2004; 92: 481-488. DOI: 10.1524/ract.92.8.481.39281
[42] Amachi S, Fujii T, Shinoyama H et al. Microbial influences on the mobility and transformation of radioactive iodine in the environment. J Nucl Radiochem Sci, 2005; 6: 21-24. DOI: 10.14494/jnrs2000.6.21
[43] Ozdemir S, Oduncu MK, Kilinc E et al. Resistance, bioaccumulation and solid phase extraction of uranium (VI) by Bacillus vallismortis and its UV-vis spectrophotometric determination. J Environ Radioact, 2017; 171: 217-225. DOI: 10.1016/j.jenvrad.2017.02.021
[44] Tsuruta T. Bioaccumulation of uranium and thorium from the solution containing both elements using various microorganisms. J Alloys Comp, 2006; 408: 1312-1315. DOI: 10.1016/j.jallcom.2005.04.131
[45] Tišáková L, Pipíška M, Godány A et al. Bioaccumulation of 137Cs and 60Co by bacteria isolated from spent nuclear fuel pools. Radioanal Nucl, 2013; 295: 737-748. DOI: 10.1007/s10967-012-1932-6
[46] Jorgensen E. Ecotoxicol. Academic press: California, USA, 2010.
[47] Keasling JD, Hupf GA. Genetic manipulation of polyphosphate metabolism affects cadmium tolerance in Escherichia coli. Appl Environ Microb, 1996; 62: 743-746. DOI: 10.1128/aem.62.2.743-746.1996
[48] Jiang W, Saxena A, Song B et al. Elucidation of functional groups on gram-positive and gram-negative bacterial surfaces using infrared spectroscopy. Langmuir, 2004; 20: 11433-11442. DOI: 10.1021/la049043
[49] Francis AJ, Nancharaiah YV. In situ and ex situ bioremediation of radionuclide-contaminated soils at nuclear and norm sites. Environ Remediat Restor Contam Nucl, 2015; 185-236. DOI: 10.1016/B978-1-78242-231-0.00009-0
[50] Hazen TC, Tabak HH. Developments in bioremediation of soils and sediments polluted with metals and radionuclides: 2. Field research on bioremediation of metals and radionuclides. Rev Environ Sci Biotechnol, 2005; 4: 157-183. DOI: 10.1007/s11157-005-2170-y
[51] Lloyd JR, Lovley DR. Microbial detoxification of metals and radionuclides. Curr Opin Biotechnol, 2001; 12: 248-253. DOI: 10.1016/S0958-1669(00)00207-X
[52] Roh C, Kang CK, Lloyd JR. Microbial bioremediation processes for radioactive waste. Korean J Chem Eng, 2015; 32: 1720-1726. DOI: 10.1007/s11814-015-0128-5
[53] Shukla A, Parmar P, Saraf M. Radiation, radionuclides and bacteria: An in-perspective review. J Environ Radioact, 2017; 180: 27-35. DOI: 10.1016/j.jenvrad.2017.09.013
[54] Gadd GM. Influence of microorganisms on the environmental fate of radionuclides. Endeavour, 1996; 20: 150-156. DOI: 10.1016/S0160-9327(96)10021-1
[55] Prakash D, Gabani P, Chandel AK et al. Bioremediation: a genuine technology to remediate radionuclides from the environment. Microb Biotechnol, 2013; 6: 349-360. DOI: 10.1111/1751-7915.12059
[56] Francis AJ, Dodge CJ. Microbial transformation of actinides and other radionuclides. The 9th Biennial daebrns. Symposium on Nuclear and Radio-chemistry, Mumbai, India, 7-10 January, 2009.
[57] Merroun ML, Selenska-Pobell S. Bacterial interactions with uranium: An environmental perspective. J Contam Hydrol, 2008; 102: 285-295. DOI: 10.1016/j.jconhyd.2008.09.019
[58] Lovley DR. Microbial redox interactions with uranium: An environmental perspective. Radioact Environ, 2002; 2: 205-223. DOI: 10.1016/S1569-4860(02)80036-5
[59] Yin Y, Wang J, Yang X et al. Removal of strontium ions by immobilized Saccharomyces cerevisiae in magnetic chitosan microspheres. Nucl Eng Technol, 2017; 49: 172-177. DOI: 10.1016/j.net.2016.09.002
[60] Suzuki Y, Banfield JF. Resistance to, and accumulation of, uranium by bacteria from a uranium-contaminated site. Geomicrobiol J, 2004; 21: 113-121. DOI: 10.1080/01490450490266361
[61] Sheppard S, Long J, Sanipelli B. Solid/liquid partition coefficients (Kd) for selected soils and sediments at Forsmark and Laxemarsimpevarp. Accessed March 1, 2009. Available at https://inis.iaea.org/search/search.aspx?orig_q=RN:40109502
[62] Özdemir S, Oduncu MK, Kilinc E et al. Tolerance and bioaccumulation of U (VI) by Bacillus mojavensis and its solid phase preconcentration by Bacillus mojavensis immobilized multiwalled carbon nanotube. J Environ Manage, 2017; 187: 490-496. DOI: 10.1016/j.jenvman.2016.11.004
[63] Amachi S, Kasahara M, Hanada S et al. Microbial participation in iodine volatilization from soils. Environ Sci Technol, 2003; 37: 3885-3890. DOI: 10.1021/es0210751
[64] Alexandrova M, Rozhko T, Vydryakova G et al. Effect of americium-241 on luminous bacteria. Role of peroxides. J Environ Radioact, 2011; 102: 407-411. DOI: 10.1016/j.jenvrad.2011.02.011
[65] Groudev SN, Georgiev PS, Spasova II et al. Bioremediation of a soil contaminated with radioactive elements. Hydrometallurgy, 2001; 59: 311-318. DOI: 10.1016/S0304-386X(00)00187-0
[66] De Filippis LF. Role of phytoremediation in radioactive waste treatment. Soil Remed Plant Prospec Challeng, 2015; 1: 207-254. DOI: 10.1016/B978-0-12-799937-1.00008-5
[67] de Mello-Farias PC, Chaves ALS, Lencina CL. Transgenic plants for enhanced phytoremediation-Physiological studies. Genet Transform, 2011; 305-328. DOI: 10.5772/24355
[68] Turnau K, Orlowska E, Ryszka P et al. Role of mycorrhizal fungi in phytoremediation and toxicity monitoring of heavy metal rich industrial wastes in southern Poland. Soil Water Pollut Prot Remediation, 2006; 533-551.
[69] Dushenkov S. Trends in phytoremediation of radionuclides. Plant Soil, 2003; 249: 167-175. DOI: 10.1023/A:1022527207359
[70] De Filippis LF. Biochemical and molecular aspects in phytoremediation of selenium. Plant Adapt Phytoremediat, 2010; 193-226. DOI: 10.1007/978-90-481-9370-7_10
[71] Sharma S, Singh B, Manchanda VK. Phytoremediation: role of terrestrial plants and aquatic macrophytes in the remediation of radionuclides and heavy metal contaminated soil and water. Environ Sci Pollut Res, 2015; 22: 946-962. DOI: 10.1007/s11356-014-3635-8
[72] Garten Jr CT, Lomax RD. Technetium-99 cycle in maple trees: Characterization of changes in chemical form. Health Phys, 1989; 57: 299-307. DOI: 10.1097/00004032-198908000-00008
[73] Garten CT, Tucker CS, Walton BT. Environmental fate and distribution of technetium-99 in a deciduous forest ecosystem. J Environ Radioact, 1986; 3: 163-188. DOI: 10.1016/0265-931X(86)90024-X
[74] Echevarria G, Vong PC, Morel JL. Bioavailability of Technetium-99 as affected by plant species and growth, application form, and soil incubation. J Environ Qual, 1997; 26: 947-956. DOI: 10.2134/jeq1997.00472425002600040004x
[75] Entry JA, Rygiewicz PT, Emmingham WH. Accumulation of cesium 137 and strontium 90 in Ponderosa pine and Monterey pine seedlings. J Environ Qual, 1993; 22: 742-746. DOI: 10.2134/jeq1993.00472425002200040016x
[76] Hoseini PS, Poursafa P, Moattar F et al. Ability of phytoremediation for absorption of strontium and cesium from soils using Cannabis sativa. Int J Env Health Eng, 2012; 1: 17. DOI: 10.4103/2277-9183.96004
[77] Singh S, Thorat V, Kaushik CP et al. Potential of Chromolaena odorata for phytoremediation of 137Cs from solution and low-level nuclear waste. J Hazard Mater, 2009; 162: 743-745. DOI: 10.1016/j.jhazmat.2008.05.097
[78] Singh S, Eapen S, Thorat V et al. Phytoremediation of 137 cesium and 90strontium from solutions and low-level nuclear waste by Vetiveria zizanoides. Ecotox Environ Safe, 2008; 69: 306-311. DOI: 10.1016/j.ecoenv.2006.12.004
[79] Eapen S, Singh S, Thorat V et al. Phytoremediation of radiostrontium (90Sr) and radiocesium (137Cs) using giant milky weed (Calotropis gigantean R.Br.) plants. Chemosphere, 2006; 65: 2071-2073. DOI: 10.1016/j.chemosphere.2006.06.049
[80] Zehnder HJ, Kopp P, Eikenberg J et al. Uptake and transport of radioactive cesium and strontium into grapevines after leaf contamination. Radiat Phys Chem, 1995; 46: 61-69. DOI: 10.1016/0969-806X(94)00115-Z
[81] Dutton MV, Humphreys PN. Assessing the potential of short rotation coppice (Src) for cleanup of radionuclide contaminated sites. Int J Phytoremediat, 2005; 7: 279-293. DOI: 10.1080/16226510500327137
[82] Entry JA, Watrud LS, Reeves M. Accumulation of 137Cs and 90Sr from contaminated soil by three grass species inoculated with mycorrhizal fungi. Environ Pollut, 1999; 104: 449-457. DOI: 10.1016/S0269-7491(98)00163-8
[83] Veresoglou DS, Barbayiannis N, Matsi T. Shoot Sr concentrations in relation to shoot Ca concentrations and to soil properties. Plant Soil, 1996; 1978: 178. DOI: 10.1007/BF00011167
[84] Dushenkov S, Vasudev D, Kapulnik Y et al. Removal of uranium from water using terrestrial plants. Environ Sci Technol, 1997; 31: 3468-3474. DOI: 10.1021/es970220l
[85] Prasad MNV. Sunflower (Helinathus annuus L.)-a potential crop for environmental industry. HELIA, 2007; 30: 167-174. DOI: 10.2298/HEL0746167P
[86] Prasad MNV, de Oliveira Freitas HM. Metal hyperaccumulation in plants-Biodiversity prospecting for phytoremediation technology. Electron J Biotech, 2003; 6: 285-321. DOI: 10.2225/vol6-issue3-fulltext-6
[87] Singh A, Eapen S, Fulekar MH. Phytoremediation technology for remediation of radiostrontium (90Sr) and radiocaesium (137Cs) by Catharanthus roseus (l.) G. Don in aquatic environment. Environ Eng Manag J, 2009; 8: 527-532. DOI: 10.30638/eemj.2009.072
[88] Rogers RD, Williams SE. Vesicular-arbuscular mycorrhiza: Influence on plant uptake of cesium and cobalt. Soil Biol Biochem, 1986; 18: 371-376. DOI: 10.1016/0038-0717(86)90040-4
[89] Lewis BG, MacDonnell MM. Release of radon-222 by vascular plants: Effect of transpiration and leaf area. J Environ Qual, 1990; 19: 93-97. DOI: 10.2134/jeq1990.00472425001900010012x
[90] Ebbs SD, Brady DJ, Kochian LV. Role of uranium speciation in the uptake and translocation of uranium by plants. J Exp Bot, 1998; 49: 1183-1190. DOI: 10.1093/jxb/49.324.1183
[91] Huang JW, Blaylock MJ, Kapulnik Y et al. Phytoremediation of uranium contaminated soils: Role of organic acids in triggering uranium hyperaccumulation in plants. Environ Sci Technol, 1998; 32: 2004-2008. DOI: 10.1021/es971027u
[92] Eapen S, Suseelan KN, Tivarekar S et al. Potential for rhizofiltration of uranium using hairy root cultures of Brassica juncea and Chenopodium amaranticolor. Environ Res, 2003; 91: 127-133. DOI: 10.1016/S0013-9351(02)00018-X
[93] Noshkin VE. Ecological aspects of plutonium dissemination in aquatic environments. Health Phys, 1972; 22: 537-549. DOI: 10.1097/00004032-197206000-00001
[94] Cataldo DA, McFadde KM, Garland TR et al. Organic constituents and complexation of Nickel (II), Iron (III), Cadmium (II), and Plutonium (IV) in soybean xylem exudates. Plant Physiol, 1998; 86: 734-739. DOI: 10.1104/pp.86.3.734
[95] Garten CT, Tucker CS, Scott TG. Plant uptake of Np-237 and Tc-99 under field conditions. J Environ Radioact, 1986; 4: 91-99. DOI: 10.1016/0265-931X(86)90035-4
[96] Kovacs T, Bodrogi E, Somlai J et al. 210Po and 210Pb-determination in Hungarian grow tobacco. AARMS, 2004; 3: 165-169.
[97] Goncharova NV, Availability of radiocesium in plant from soil: Facts, mechanisms andmodelling. Global Nest J, 2009; 11: 260-266. DOI: 10.30955/gnj.000609
[98] Chou FI, Chung HP, Teng SP et al. Screening plant species native to Taiwan for remediation of 137Cs-contaminated soil and the effects of K addition and soil amendment on the transfer of 137Cs from soil to plants. J Environ Radioactiv, 2005; 80: 175-181. DOI: 10.1016/j.jenvrad.2004.10.002
[99] Salt DE, Pickering IJ, Prince RC et al. Metal accumulation by aquacultured seedlings of Indian mustard. Environ Sci Technol. 1997; 31: 1636-1644. DOI: 10.1021/es960802n
[100] Tkavc R, Matrosova VY, Grichenko OE et al. Prospects for fungal bioremediation of acidic radioactive waste sites: characterization and genome sequence of Rhodotorulataiwanensis MD1149. F Microbio, 2018; 8: 2528. DOI: 10.3389/fmicb.2017.02528
[101] Singh M, Singh D, Rai PK et al. Fungi in remediation of hazardous wastes: Current status and future outlook. Recent Trends Mycol Res, 2021; 195-224. DOI: 10.1007/978-3-030-68260-6_8
[102] Ehrlich HL, Newman DK. Geomicrobiology, 5th ed. CRC Press: New York: USA, 2008. DOI: 10.1201/9780849379079
[103] Culotta VC, Daly MJ. Manganese complexes: diverse metabolic routes to oxidative stress resistance in prokaryotes and yeast. Antioxid Redox Sign, 2013; 19: 933-944. DOI: 10.1089/ars.2012.5093
[104] Galanda D, Mátel Ľ, Strišovská J et al. Mycoremediation: The study of transfer factor for plutonium and americium uptake from the ground. J Radioanal Nucl Chem, 2014; 299: 1411-1416. DOI: 10.1007/s10967-013-2909-9
[105] Das N. Remediation of radionuclide pollutants through biosorption-an overview. Clean Soil Air Water, 2012; 40: 16-23. DOI: 10.1002/clen.201000522
[106] Dhami PS, Gopalakrishnan V, Kannan R et al. Biosorption of Radionuclides by Rhizopus arrhizus. Biotechnol Lett, 1998; 20: 225. DOI: 10.1023/A:1005313532334
[107] Akhtar K, Akhtar MW, Khalid AM. Removal and recovery of uranium from aqueous solutions by Trichoderma harzianum. Water Res, 2007; 41: 1366. DOI: 10.1016/j.watres.2006.12.009
[108] Fomina MA, Kadoshnikov VM, Zlobenko BP. Fungal biomass grown on media containing clay as a sorbent of radionuclides. Process Biochem, 1999; 9: 245. DOI: 10.1016/S1572-4409(99)80114-X
[109] Dhami PS, Kannan R, Naik PW et al. Biosorption of americium using various biomasses of Rhizopus species. Biotechnol Lett, 2002; 24: 885-889. DOI: 10.1023/A:1015533129642
[110] Roy K, Sinha P, Lahiri S. Immobilization of long-lived radionuclides Eu152,154 by selective bioaccumulation in saccharomyces cerevisiae from a synthetic mixture of Eu152,154, 137Cs and 60Co. Biochem Eng J, 2008; 40: 363-367. DOI: 10.1016/j.bej.2008.01.005
[111] Bhatti HN, Farooqi ZH. Modern physical chemistry. Caravan Book House Lahore: Lahore, Pakistan, 2013.
[112] Poinssot C, Geckeis H. Radionuclide behaviour in the natural environment: science, implications and lessons for the nuclear industry. Elsevier: New York, USA, 2012. DOI: 10.1533/9780857097194
[113] Payne RB, Gentry DM, Rapp-Giles BJ et al. Uranium reduction by Desulfovibriode sulfuricans strain G20 and a cytochrome c3 mutant. Appl Environ Microb, 2002; 68: 3129-3132. DOI: 10.1128/AEM.68.6.3129-3132.2002
[114] Tsuruta T. Bioaccumulation of uranium and thorium from the solution containing both elements using various microorganisms. J Alloys Comp, 2006; 408: 1312-1315. DOI: 10.1016/j.jallcom.2005.04.131
[115] Lovley DR, Phillips EJ. Reduction of uranium by Desulfovibriode sulfuricans. Appl Environ Microb, 1992; 58: 850-856. DOI: 10.1128/aem.58.3.850-856.1992
[116] Beliaev AS, Klingeman DM, Klappenbach JA et al. Global transcriptome analysis of Shewanellaoneidensis MR-1 exposed to different terminal electron acceptors. J Bacteriol, 2005; 187: 7138-7145. DOI: 10.1128/JB.187.20.7138-7145.2005
[117] Marshall MJ, Beliaev AS, Dohnalkova AC et al. C-Type cytochrome-dependent formation of U(IV) nanoparticles by Shewanel laoneidensis. PLoS biology, 2006; 4: e268. DOI: 10.1371/journal.pbio.0040268
[118] Lovley DR, Phillips EJP, Gorby YA et al. Microbial reduction of uranium. Nature, 1991; 350: 413-416. DOI: 10.1038/350413a0
[119] Orellana R, Leavitt JJ, Comolli LR et al. U (VI) reduction by diverse outer surface c-type cytochromes of Geobacter sulfurreducens. Appl Environ Microb, 2013; 79: 6369-6374. DOI: 10.1128/AEM.02551-13
[120] Shi L, Chen B, Wang Z et al. Isolation of a high-affinity functional protein complex between OmcA and MtrC: Two outer membrane decaheme c-type cytochromes of Shewanellaoneidensis MR-1. J Bacteriol, 2006; 188: 4705-4714. DOI: 10.1128/JB.01966-05
[121] Hartshorne RS, Reardon CL, Ross D et al. Characterization of an electron conduit between bacteria and the extracellular environment. P Natl Acad Sci, 2009; 106: 22169-22174. DOI: 10.1073/pnas.0900086106
[122] Fisher NS, Bjerregaard P, Huynh-Ngoc L et al. Interactions of marine plankton with transuranic elements. II. Influence of dissolved organic compounds on americium and plutonium accumulation in a diatom. Mar Chem, 1983; 13: 45-56. DOI: 10.1016/0304-4203(83)90048-8
[123] Kaszuba JP, Runde WH. The aqueous geochemistry of neptunium: Dynamic control of soluble concentrations with applications to nuclear waste disposal. Environ Sci Technol, 1999; 33: 4427-4433. DOI: 10.1021/es990470x
[124] Songkasiri W, Reed DT, Rittmann BE. Bio-sorption of neptunium (V) by Pseudomonas fluorescens. Radiochim Acta, 2002; 90: 785-789. DOI: 10.1524/ract.2002.90.9-11_2002.785
[125] Strandberg GW, Arnold Jr WD. Microbial accumulation of neptunium. J Ind Microb, 1988; 3: 329-331. DOI: 10.1007/BF01569534
[126] Rittmann BE, Banaszak JE, Reed DT. Reduction of Np (V) and precipitation of Np (IV) by an anaerobic microbial consortium. Biodegradation, 2002; 13: 329-342. DOI: 10.1023/A:1022382627690
[127] Lloyd JR, Chesnes J, Glasauer S et al. Reduction of actinides and fission products by Fe (III)-reducing bacteria. Geomicrobiol J, 2002; 19: 103-120. DOI: 10.1080/014904502317246200
[128] Rusin PA, Quintana L, Brainard JR et al. Solubilization of plutonium hydrous oxide by iron-reducing bacteria. Environ Sci Technol, 1994; 28: 1686-1690. DOI: 10.1021/es00058a021
[129] Neu MP, Icopini GA, Boukhalfa H. Plutonium speciation affected by environmental bacteria. Radiochim Acta, 2005; 93: 705-714. DOI: 10.1080/01490450701457030
[130] Kimber RL, Boothman C, Purdie P. Biogeochemical behavior of plutonium during anoxic biostimulation of contaminated sediments. Mineral Mag, 2012; 76: 567-578. DOI: 10.1180/minmag.2012.076.3.08
[131] de Pádua Ferreira RV, Sakata SK et al. Influence of americium-241 on the microbial population and biodegradation of organic waste. Environ Chem Lett, 2011; 9: 209-216. DOI: 10.1007/s10311-009-0266-2
[132] Bondietti EA, Francis CW. Geologic migration potentials of technetium-99 and neptunium-237. Science, 1979; 203: 1337-1340. DOI: 10.1126/science.203.4387.1337
[133] Burke IT, Boothman C, Lloyd JR et al. Reoxidation behavior of technetium, iron, and sulfur in estuarine sediments. Environ Sci Technol, 2006; 40: 3529-3535. DOI: 10.1021/es052184t
[134] McBeth JM, Lear G, Lloyd JR et al. Technetium reduction and reoxidation in aquifer sediments. Geomicrobiol J, 2007 24: 189-197. DOI: 10.1080/01490450701457030
[135] Lloyd JR, Ridley J, Khizniak T et al. Reduction of technetium by Desulfovibriodesulfuricans: Biocatalyst characterization and use in a flow through bioreactor. Appl Environ Microb, 1999; 65: 2691-2696. DOI: 10.1128/AEM.65.6.2691-2696.1999
[136] Lloyd JR, Yong P, Macaskie LE. Biological reduction and removal of Np (V) by two microorganisms. Environ Sci Technol, 2000; 34: 1297-1301. DOI: 10.1021/es990394y
[137] Fauque GD. Ecology of sulfate-reducing bacteria. Springer: Massachusetts, USA, 1995. DOI: 10.1007/978-1-4899-1582-5_8
[138] Lloyd JR, Cole JA, Macaskie LE. Reduction and removal of heptavalent technetium from solution by Escherichia coli. J Bacteriol, 1997; 179: 2014-2021. DOI: 10.1128/jb.179.6.2014-2021.1997
[139] De Luca G, de Philip P, Dermoun Z et al. Reduction of technetium (VII) by Desulfovibrio fructosovorans is mediated by the nickel-iron hydrogenase. Appl Environ Microb, 2001; 67: 4583-4587. DOI: 10.1128/AEM.67.10.4583-4587.2001
[140] Macaskie LE. The application of biotechnology to the treatment of wastes produced from the nuclear fuel cycle: biodegradation and bioaccumulation as a means of treating radionuclide-containing streams. Crit Rev Biotechnol, 1991; 11: 41-112. DOI: 10.3109/07388559109069183
[141] Faison BD, Cancel CA, Lewis SN et al. Binding of dissolved strontium by Micrococcus luteus. Appl Environ Microb, 1990; 56: 3649-3656. DOI: 10.1128/aem.56.12.3649-3656.1990
[142] Ilyas S, Srivastava RR, Ilyas N. Biosorption of Strontium from Aqueous Solutions. Strontium Contam Environ, 2020; 65-83. DOI: 10.1007/978-3-030-15314-4_4
[143] Dwivedy KK, Mathur AK. Bioleaching-Our experience. Hydrometallurgy, 1995; 38: 99-109. DOI: 10.1016/0304-386X(94)00034-Z
[144] Elbaradei M, Nwogugu E, Rames J. International law and nuclear energy: Overview of the legal framework. IAEA Bulletin, 1995; 37: 16-25.
[145] Abdel Rahman RO. Planning and implementation of radioactive waste management system. IntechOpen: London, UK, 2012. DOI: 10.5772/39056
Copyright © 2023 The Author(s). This open-access article is licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, sharing, adaptation, distribution, and reproduction in any medium, provided the original work is properly cited.




 Copyright ©
Copyright ©