Explainable Artificial Intelligence Methods for Breast Cancer Recognition
Robertas Damaševičius1*
1Department of Applied Informatics, Vytautas Magnus University, Kaunas, Lithuania
*Correspondence to: Robertas Damaševičius, PhD, Professor, Department of Applied Informatics, Vytautas Magnus University, K. Donelaičio St. 58 Kaunas, Kaunas, LT-440011, Lithuania; Email: robertas.damasevicius@vdu.lt
DOI: 10.53964/id.2024025
Abstract
Breast cancer remains a leading cause of cancer-related mortality among women worldwide, necessitating early and accurate detection for effective treatment and improved survival rates. Artificial intelligence (AI) has shown significant potential in enhancing the diagnostic and prognostic capabilities in breast cancer recognition. However, the black-box nature of many AI models poses challenges for their clinical adoption due to the lack of transparency and interpretability. Explainable AI (XAI) methods address these issues by providing human-understandable explanations of AI models’ decision-making processes, thereby increasing trust, accountability, and ethical compliance. This review explores the current state of XAI methods (Local Interpretable Model-agnostic Explanations, Shapley Additive explanations, Gradient-weighted Class Activation Mapping) in breast cancer recognition, detailing their applications in various tasks such as classification, detection, segmentation, prognosis, and biomarker discovery. By integrating domain-specific knowledge and developing visualization techniques, XAI methods enhance the usability and interpretability of AI systems in clinical settings. The study also identifies the key challenges and future directions in the evaluation of XAI methods, the development of standardized metrics, and the seamless integration of XAI into clinical workflows.
Keywords: explainable artificial intelligence, medical imaging, breast cancer recognition
1 INTRODUCTION
1.1 Background on Breast Cancer and Its Impact
Breast cancer is a significant global health concern, affecting millions of women worldwide. As the most prevalent cancer among women, its incidence rate has been on a steady rise, leading to increased mortality rates and health disparities across different regions and populations[1]. Early detection and accurate diagnosis are pivotal in improving survival rates, underscoring the necessity for the development of effective diagnostic tools[2].
The economic burden of breast cancer is substantial, encompassing both direct and indirect costs. Direct costs include expenses related to medical care, diagnosis, and treatment, such as hospitalization, surgery, chemotherapy, radiotherapy, targeted therapy, and imaging tests[3]. Indirect costs encompass productivity loss and societal impacts, including loss of income due to work absence or reduced working capacity, premature.
Mortality, years of potential life lost, and caregiver burden[4]. The global economic impact of breast cancer is immense, straining healthcare systems and affecting patients and their families[5].
Breast cancer screening and diagnosis are paramount in early detection, which contributes to improved survival rates. Mammography is the primary screening tool, while other imaging modalities like ultrasound, MRI, and tomosynthesis also play a role[6]. Screening programs have proven effective in reducing mortality rates, but challenges in breast cancer diagnosis persist. These challenges include subjectivity and variability in radiologists’ interpretations, false positives and negatives leading to unnecessary biopsies or delayed treatment, and limited access to screening and diagnostic services in low-resource settings[7].
The need for improved precision diagnostic techniques is evident[8], and artificial intelligence (AI) has the potential to transform breast cancer recognition. Deep learning models (e.g., DeepSHAP) can analyze and interpret medical images, predict treatment response and prognosis, and facilitate personalized treatment planning[9-11]. The adoption of AI-based diagnostic tools in clinical settings is hindered by issues related to transparency, interpretability, ethical considerations, regulatory hurdles, and integration into clinical workflows[12-15].
1.2 Role of Imaging Modalities in Breast Cancer Recognition
Breast cancer recognition relies on various imaging modalities such as mammography[6,16], ultrasound[9,10,17,18], magnetic resonance imaging (MRI)[19], and histopathology images[20]. Radiologists analyze these images to identify suspicious lesions and assess their malignancy. The interpretation of these images can be subjective and prone to human error. AI techniques, especially deep Shapley additive explanations (DeepSHAP), have demonstrated remarkable performance in breast cancer recognition tasks. Their lack of transparency and interpretability can limit their clinical utility. The explainable AI (XAI) methods have emerged as a promising approach to address these limitations, providing an understanding of the AI models’ decision-making process and enabling more informed clinical decisions[21,22].
Imaging modalities play a crucial role in breast cancer recognition, as they enable the early detection, diagnosis, and monitoring of the disease. Various imaging techniques are used to visualize breast tissue, identify suspicious lesions, and assess their malignancy. The choice of the appropriate imaging modality depends on factors such as the patient’s age, breast density, and risk factors for breast cancer. The main imaging modalities used in breast cancer recognition include mammography, ultrasound, MRI, and digital breast tomosynthesis (DBT).
Mammography[23] is the gold standard for breast cancer screening and is recommended for women of average risk starting at age 40 or 50, depending on the guidelines followed. It uses low-dose X-rays to produce images of breast tissue, which can reveal calcifications, masses, and architectural distortions that may indicate the presence of breast cancer. Mammography is highly effective in detecting breast cancer in its early stages, which significantly improves treatment outcomes and survival rates. However, mammography has some limitations, including reduced sensitivity in dense breast tissue, false positives, and ionizing radiation exposure.
Breast ultrasound[24,25] uses high-frequency sound waves to create images of breast tissue, making it a radiation-free modality. Ultrasound is often used as an adjunct to mammography, especially in women with dense breasts or those who cannot undergo.
It can differentiate between solid masses and fluid-filled cysts and guide biopsies of suspicious lesions. While ultrasound is a valuable supplementary tool, it has lower specificity than mammography, which may result in more false-positive findings.
Breast MRI[26] is a highly sensitive imaging modality that uses a powerful magnetic field and radio waves to generate detailed images of breast tissue. MRI is recommended for women with a high risk of breast cancer, such as those with a strong family history or a known genetic mutation like Breast invasive Carcinoma (BRCA)1 or BRCA2[27]. It is also used for evaluating the extent of the disease in newly diagnosed patients and monitoring response to neoadjuvant therapy. Although MRI has high sensitivity, it is associated with a higher rate of false positives and is more expensive than other imaging techniques.
DBT, also known as 3D mammography, is a relatively new imaging modality that acquires multiple low-dose X-ray images at different angles, which are then reconstructed into a three-dimensional representation of the breast[28,29]. DBT improves the detection of breast cancer, particularly in women with dense breasts, by reducing the overlapping of breast tissue seen in conventional mammography. This technology has been shown to increase cancer detection rates and reduce the number of false positives and unnecessary biopsies[30].
The role of imaging modalities in breast cancer recognition is vital, as they facilitate early detection, diagnosis, and monitoring of the disease. Each modality has its advantages and limitations, and their combined use can provide a comprehensive assessment of breast tissue, improving the accuracy of breast cancer recognition and ultimately enhancing patient care.
1.3 Potential of AI in Breast Cancer Diagnosis and Prediction
AI techniques, particularly those involving deep learning and machine learning algorithms, have demonstrated significant potential in breast cancer diagnosis and prediction[29,31,32]. These techniques can analyze complex medical data, identify patterns, and make predictions that can assist healthcare professionals in making more accurate and timely decisions[33]. The potential of AI techniques in breast cancer diagnosis and prediction can be explored through various aspects, including image analysis, classification, treatment response prediction, and personalized medicine[34-36].
AI techniques can enhance the analysis of medical images acquired through different imaging modalities, such as mammography[37], ultrasound, MRI, and DBT. DeepSHAP, such as convolutional neural networks (CNNs), have shown exceptional performance in detecting and classifying breast lesions. These models can identify subtle features and patterns that may be overlooked by human observers, leading to improved diagnostic accuracy and reduced interobserver variability[38].
The AI algorithms can be trained to classify breast lesions as benign or malignant and assess the risk of developing breast cancer[39]. These classifications can be based on a combination of imaging features, patient demographics, and clinical history. By providing accurate and consistent classifications, AI techniques can help reduce the number of unnecessary biopsies, lower false-positive rates, and minimize anxiety for patients.
AI techniques can be employed to predict the response to various breast cancer treatments, such as chemotherapy, radiotherapy, and hormone therapy[36]. By analyzing imaging, genomic, and clinical data, machine learning models can identify biomarkers and patterns associated with treatment response[40]. This information can help health.
Care professionals make informed decisions on treatment planning, improving patient outcomes and reducing the likelihood of over- or under-treatment.
Breast cancer is a heterogeneous disease with diverse molecular subtypes, clinical behavior, and prognosis. Genomic and transcriptomic profiling has become instrumental in advancing the diagnosis and treatment of breast cancer, offering a molecular-level understanding of individual tumors. This approach enables the identification of genetic alterations and gene expression patterns that drive the disease, facilitating personalized medicine strategies that are tailored to the specific molecular characteristics of a patient’s tumor[41]. Genomic profiling involves analyzing the DNA sequences in breast cancer cells to identify mutations and variations. These genetic markers can help predict how aggressive the cancer is likely to be and suggest the most effective treatment options[42]. Technologies such as next-generation sequencing have revolutionized this field by allowing for rapid, comprehensive analyses of genomic alterations[43]. Transcriptomic profiling assesses the RNA expressions to understand which genes are active in breast cancer cells. This profiling provides insights into the functional consequences of genetic alterations observed in the genomic profile. It helps in understanding the tumor environment, predicting response to specific treatments, and identifying potential resistance mechanisms to existing therapies[44]. The integration of genomic and transcriptomic data has led to the development of targeted therapies and improved prognostic models, significantly enhancing patient outcomes[45]. AI techniques can analyze large-scale data from sources such as gene expression profiles, genomic alterations, and clinical information to identify specific patterns and subtypes[46]. This information can be used to predict patient outcomes, such as recurrence and survival rates, and guide personalized treatment plans tailored to each patient’s unique characteristics.
Despite the promising potential of AI techniques in breast cancer diagnosis and prediction[47], several challenges remain, including the need for high-quality and diverse data, the integration of AI tools into clinical workflows, and the “black-box” nature of some AI models. Addressing these challenges, particularly by improving model transparency and interpretability through XAI methods, will be essential to fully harness the potential of AI techniques in breast cancer diagnosis and prediction and improve patient outcomes[48].
1.4 Objectives and Contributions
This review aims to explore the potential and challenges of XAI methods in breast cancer recognition, providing a comprehensive review of recent advances and identifying future research directions. More specifically, the objectives of this study are to: (1) detail the challenges associated with the black-box nature of AI models, exploring the implications for trust, accountability, ethical considerations, and model improvement. (2) Review the methodologies and applications of key XAI methods in breast cancer recognition, including local interpretable model-agnostic explanations (LIME), Shapley additive explanations (SHAP), and gradient-weighted class activation mapping (Grad-CAM). (3) Discuss the challenges and future directions in the evaluation of explanations, development of domain-specific XAI techniques, and integration of XAI methods into the clinical workflow.
Through these objectives, this study intends to provide valuable insights into the promising field of XAI in breast cancer recognition, contributing to the ongoing research and development efforts aimed at enhancing the clinical utility and interpretability of AI-based diagnostic tools.
The contributions of this paper are: (1) Present an overview of XAI in breast cancer recognition, capturing the latest research advancements, methodologies, and applications. (2) Identify current challenges and point out promising future research avenues, potentially guiding the development and evaluation of XAI methods in the future. (3) Contribute to the broader discourse on the interpretability of AI systems, fostering a deeper understanding and broader adoption of AI in healthcare settings, and potentially leading to improved patient outcomes.
2 METHODS
Several XAI methods have been employed to improve the interpretability of AI models for breast cancer recognition. Some prominent methods include LIME, SHAP, and Grad-CAM. These are explained in further sections in more detail.
2.1 Black-box Nature of AI Models and The Need for XAI
The black-box nature of AI models, particularly DeepSHAP such as CNNs, refers to the limited transparency and interpretability of these models’ decision-making processes[49]. Although AI models have shown remarkable performance in various applications, including breast cancer diagnosis and prediction, understanding the rationale behind their predictions is challenging. This lack of transparency can hinder the adoption of AI models in clinical settings, where trust and accountability are paramount. XAI has emerged as a solution to address the black-box problem and enhance the interpretability of AI models[13,50].
The black-box nature of AI models poses several challenges, especially in the context of healthcare[51]: (1) Clinicians may be reluctant to rely on AI models for decision-making if they cannot understand or validate the reasoning behind the predictions. Trust is essential for the adoption of AI tools in clinical practice. (2) In healthcare, decision-makers must be accountable for their actions. The lack of transparency in AI models can make it difficult to assign responsibility in cases where errors or adverse outcomes occur. (3) AI models must adhere to ethical principles, such as fairness, transparency, and non-maleficence. Regulatory bodies also require evidence of the safety and effectiveness of AI tools, which may be difficult to demonstrate if the models’ decision-making processes are not transparent. (4) Without insights into the decision-making process, it can be challenging to identify the limitations of AI models and make improvements, potentially leading to suboptimal performance or biased predictions.
XAI aims to provide transparency and interpretability in AI models’ decision-making processes, addressing the challenges posed by the black-box nature of these models. By generating human-understandable explanations for the predictions, XAI methods can help clinicians and other stakeholders build trust in AI models, ensure accountability, meet ethical and regulatory requirements, and facilitate model improvement. Key aspects of XAI include: (1) Local explanations: Providing explanations for individual predictions or decisions, helping clinicians understand the specific reasoning behind each case[53]. (2) Global explanations: Offering insights into the general decision-making process of the AI model, which can help users understand how the model behaves across different cases and identify potential biases or limitations. (3) Visualization techniques: Presenting the explanations in an intuitive and easily understandable manner, such as highlighting important features or regions in medical images that the AI model has based its decision on.
The black-box nature of AI models poses significant challenges in the context of healthcare, where trust, accountability, and transparency are crucial[54,55]. XAI methods can address these challenges by enhancing the interpretability of AI models, fostering trust, and enabling their successful adoption in clinical settings, ultimately leading to improved patient outcomes.
2.2 LIME
LIME is a model-agnostic method that provides local explanations for individual predictions of machine learning models[56]. The key idea of LIME is to approximate the complex model locally around the prediction to be explained by a simpler, interpretable model. It generates a set of perturbed instances around the input and trains an interpretable model, such as a linear model or decision tree, to approximate the original model’s behavior locally. LIME has been employed to explain the predictions of AI models in breast cancer classification, helping radiologists understand the rationale behind the model’s decisions. This method has been applied to a variety of breast cancer recognition tasks, including classification and segmentation, offering valuable insights into the decision-making process of CNNs.
The LIME procedure can be formalized as follows: Given an instance x to explain and a black-box model f, the first step is to generate a set of perturbed instances x′ around x and compute their corresponding predictions f(x′). The next step is to compute the similarity π(x, x′) between the original instance x and each perturbed instance x′, which measures how close they are. Finally, an interpretable model g (e.g., linear or decision tree model) is fitted on the perturbed instances, using the predictions f(x′) as the target and the similarity π(x, x′) as weights.
The fitting of the interpretable model can be defined as the solution of the following.
$$ \underset{{g}\in {G}}{{min}}\sum _{x{\text{'}},z{\text{'}}}\pi \left(x,x{\text{'}}\right){\left(f\left(x{\text{'}}\right)-g\left(z{\text{'}}\right)\right)}^{2}+{\Omega }\left({g}\right)\left(1\right)$$
where G is the class of interpretable models, z′ is the interpretable representation of the perturbed instance x′ (e.g., binary vector indicating the presence or absence of words in text data, or superpixels in image data), f(x′) is the prediction of the black-box model for x′, π(x, x′) is the similarity between x and x′ , and Ω(g) is a measure of the complexity of the interpretable model g (e.g., the number of non-zero coefficients in a linear model).
The interpretable model g provides an explanation for the prediction at instance x, showing which features in the interpretable representation z are important for the prediction. Despite its simplicity, LIME can provide valuable insights into the behavior of complex models and help detect potential mistakes or biases.
In the context of breast cancer recognition tasks, LIME can be used to identify which features in a mammogram or histopathological image the CNN is focusing on to distinguish between benign and malignant tumors or to segment the tumor from the surrounding tissue. In the task of breast cancer classification, LIME can provide a visual explanation of the model’s decision-making process. By highlighting the regions in the image that the model considers significant for its prediction, LIME offers a form of validation for the model’s predictions. This can assist clinicians in understanding the basis of the model’s decision, potentially aiding in their final diagnosis. In the task of breast cancer segmentation, LIME can be used to identify the features that the model consid-ers to be part of the tumor. This can be particularly useful in cases where the tumor boundaries are not clear, providing a visual guide that can assist clinicians in determining the extent of the tumor. The application of LIME in these tasks not only enhances the interpretability of DeepSHAP but also builds trust in their predictions. By providing visual explanations for their decisions, these models become less of a “black box”, and their predictions can be validated against the expert knowledge of clinicians. This is particularly important in the medical field, where the stakes are high and the predictions of these models can have.
In the context of breast cancer recognition tasks, LIME can be used in various ways: LIME can help in understanding which features of a mammogram or histopathological image were most relevant in classifying a tumor as benign or malignant. This can provide valuable insights to radiologists and pathologists, potentially improving diagnostic accuracy. In tasks where the goal is to segment or outline the tumor in an image, LIME can provide a heatmap that highlights the areas of the image that were most relevant in identifying the tumor. This can help in evaluating the accuracy of the segmentation and in understanding the model’s decision-making process. Enhancing interpretability: One of the main challenges with machine learning models is their “black box” nature, meaning it’s often difficult to understand how they make their decisions. By providing explanations for each prediction, LIME can make these models more interpretable, leading to greater trust in their predictions.
2.3 SHAP
SHAP is a unified measure of feature importance that assigns each feature an importance value for a particular prediction[57]. Its name and theory are derived from the concept of Shapley values in cooperative game theory. Shapley values provide a fair distribution of the total payoff of a game to its players based on their contribution. In the context of machine learning, the “game” is the prediction task, the “players” are the features, and the “payoff” is the prediction. SHAP assigns each feature an importance value for a particular prediction, which is the average marginal contribution of that feature across all possible feature subsets. This ensures that the sum of the SHAP values for all features equals the difference between the prediction and the average prediction for all instances.
Given a game with n players, where each player corresponds to a feature in the model, a coalition S of players is a subset of all players. The value of a coalition, v(S), is defined as the prediction of the model with features in S turned on, minus the prediction of the model with all features turned off. The Shapley value ϕi for player i is then defined as:
$$ {{\varnothing }}_{{i}}=\sum _{S\subseteq N\setminus \left\{i\right\}}\frac{\left|{S}\right|!({n}-|{S}|-1)!}{n!}\left[v\left(S\cup \left\{i\right\}\right)-v\left(S\right)\right]\left(2\right)$$
where N is the set of all players, |S| is the number of players in S, and n is the total number of players. The term |S|!(n−|S|−1)! is the weight for each coalition, which is chosen. n! such that all permutations of players are equally likely.
SHAP has several desirable properties. It is the only method that satisfies efficiency (the feature importances sum up to the total importance), symmetry (identical features get identical importances), dummy (a feature that does not improve the prediction gets no importance), and additivity (for ensemble models, the feature importances sum up to the total importance). SHAP provides both local interpretability (explaining individual predictions) and global interpretability (explaining the whole model by averaging the SHAP values of all instances). It is model-agnostic and can be used with any model, although specific efficient algorithms are available for tree-based models and DeepSHAP. Despite its advantages, SHAP can be computationally intensive, especially for models with a large number of features or complex interactions. The interpretation of SHAP values can be challenging due to highly correlated features.
SHAP is a powerful tool for interpreting machine learning models, and its application in breast cancer recognition tasks has been demonstrated in several studies. For instance, Zhang[58] used SHAP to analyze the important factors affecting the prognosis of breast cancer patients. The study found that tumor stage, TNM stage, grade, and age have a significant impact on the prognosis of breast cancer patients. In another study, Zhao and Jiang[59] developed a machine learning model using the SHAP framework to predict distant metastasis in male breast cancer patients. The study found that the model using SHAP had the best predictive effect among all the models tested. Çubuk et al.[60] used SHAP in combination with Gaussian Processes to model both metabolic and signaling pathway activities of BRCA. They found that several metabolites have a strong impact on signaling circuits, pointing to a complex crosstalk between signaling and metabolic pathways. Mendonca-Neto et al.[61] used SHAP values for gene analysis in the classification of breast cancer subtypes. They found that certain genes are important for the classification of each subtype. These studies demonstrate the potential of SHAP in providing interpretable insights into the complex factors influencing breast cancer recognition tasks.
2.4 Grad-CAM
Grad-CAM is a visualization technique that highlights the input regions most responsible for a specific class prediction[62]. It computes the gradients of the class score with respect to the feature map activations, producing a coarse localization map that highlights the discriminative regions. Grad-CAM has been utilized in various breast cancer recog-nition tasks, such as lesion segmentation and malignancy prediction, to provide visual explanations for the model’s predictions. The idea behind Grad-CAM is to first compute.
The gradients of the output score for a particular class with respect to the feature maps of the final convolutional layer. These gradients serve as weights indicating the importance of each feature map in making the final decision. The feature maps are then combined into a single map by performing a weighted sum using these gradients, followed by a ReLU activation to keep only the positive influences. Formally, the computation of the Grad-CAM heatmap L can be defined as follows:
$$ {L}_{{G}{r}{а}{d}-{C}{A}{M}}^{c}=\left(ReLU\sum _{k}{\alpha }_{k}^{c}{A}^{k}\right)\left(3\right)$$
where LcGrad - CAM is the Grad-CAM heatmap for class c, Ak represents the k-th feature; map, and αkc is the weight of the k-th feature map for class c. The weights αkc are computed as the gradients of the score for class c (before the softmax) with respect to the feature maps:
$$ {\alpha }_{k}^{c}=\frac{1}{Z}\sum _{i}\sum _{j}\frac{\partial {y}^{c}}{{\partial A}_{ij}^{k}}\left(4\right)$$
where yc is the score for class c, Akij is the activation of the k-th feature map at spatial location (i, j), and Z is a normalization constant, typically the number of pixels in the feature map. The resulting Grad-CAM heatmap can be overlaid on the input image to visualize which regions contributed most to the model’s decision, providing valuable insights into the model’s behavior and helping identify potential mistakes or biases.
Grad-CAM has emerged as a powerful tool for enhancing the interpretability of DeepSHAP, particularly in the domain of medical imaging[63]. This technique has been applied to various breast cancer recognition tasks, including classification and segmentation, providing valuable insights into the decision-making process of CNNs. Grad-CAM works by producing a coarse localization map of important regions in the image for a particular category. This is achieved by using the gradients of target class, inputted into the final convolutional layer to create a localization map showing influential regions in the image for predicting the target class.
In the context of breast cancer recognition tasks, Grad-CAM can highlight areas in mammograms or histopathological images that the model considers significant for making its predictions. In the task of breast cancer classification, Grad-CAM can be used to visualize which regions in a mammogram or histopathological image the CNN is focusing on to distinguish between benign and malignant tumors. This not only provides a form of validation for the model’s predictions but also offers clinicians a visual explanation of the model’s decision, potentially aiding in their final diagnosis. In the task of breast cancer segmentation, Grad-CAM can be used to highlight the regions that the model considers to be part of the tumor. This can be particularly useful in cases where the tumor boundaries are not clear, providing a visual guide that can assist clinicians in determining the extent of the tumor. The application of Grad-CAM in these tasks not only enhances the interpretability of DeepSHAP but also builds trust in their predictions. By providing visual explanations for their decisions, these models become less of a “black box”, and their predictions can be validated against the expert knowledge of clinicians. This is particularly important in the medical field, where the stakes are high and the predictions of these models can have significant implications for patient care.
2.5 Summary
The applications of XAI methods in breast cancer tasks are summarized in Table 1.
Table 1. Applications of XAI Methods in Different Breast Cancer Tasks
|
LIME |
SHAP |
Grad-CAM |
Classification |
[39,63-65] |
[66,67] |
[39,68-72] |
Detection |
[73,74] |
[67,75-77] |
[78-82] |
Segmentation |
|
[5,83] |
[80] |
Prognosis |
[63,84] |
[85-89] |
|
Biomarker Discovery |
|
[7,90,91] |
|
The XAI methods are compared in Table 2 as follows: “Interpretability” refers to how easy it is for a human to understand the explanations provided by the method. “Computation Time” refers to how long it takes for the method to generate an explanation. “Applicability” refers to the types of models that the method can be applied to. “Explanation Type” refers to whether the method provides local explanations (i.e., explanations for individual predictions) or global explanations (i.e., explanations for the model’s behavior in general), or both. “Model Dependency” refers to whether the method is model-agnostic (i.e., can be applied to any type of model) or model-specific (i.e., can only be applied to certain types of models).
Table 2. Comparison of XAI Methods According to Various Criteria
|
LIME |
SHAP |
Grad-CAM |
Interpretability |
High |
Medium |
Low |
Computation time |
Fast |
Slow |
Medium |
Applicability |
General |
General |
Convolutional networks |
Explanation type |
Local |
Both |
Both |
Model dependency |
Model-agnostic |
Model-agnostic |
Model-specific |
3 APPLICATIONS OF XAI FOR BREAST CANCER DIAGNOSTICS
XAI aims to address the interpretability and transparency issues associated with traditional AI models. In the context of breast cancer diagnostics, XAI techniques have been applied in various scenarios, providing valuable insights into the decision-making process of AI models and enhancing their trustworthiness.
3.1 Domain-specific XAI Techniques
Incorporating domain knowledge into the development and application of XAI techniques is of paramount importance. Domain knowledge can provide valuable context that can enhance the interpretability and usability of AI systems. For instance, in medical imaging, domain knowledge about the anatomy, pathology, and imaging techniques can help in generating more meaningful and clinically relevant explanations[92].
Domain knowledge can be incorporated into XAI techniques in various ways. One approach is to design AI models that can incorporate domain-specific features or rules. For instance, in medical imaging, an AI model can be designed to consider the specific.
Characteristics of different types of tissues or lesions. Another approach is to use domain knowledge to guide the generation of explanations. For example, in a game design context, explanations can be generated based on the specific rules and mechanics of the game[93].
Moreover, domain-specific XAI techniques can be developed to address the specific needs and challenges of different domains. For instance, in high-risk decision-making tasks, such as identifying edible mushrooms, XAI techniques can be designed to provide detailed explanations that can help users make safer and more informed decisions[94].
The development of domain-specific XAI techniques also poses several challenges. One challenge is how to effectively incorporate domain knowledge into AI models and explanations. This requires a deep understanding of the domain as well as the AI techniques. Another challenge is how to evaluate the effectiveness of domain-specific XAI techniques. This requires the development of domain-specific evaluation metrics and benchmarks. Domain-specific XAI techniques hold great promise for enhancing the interpretability and usability of AI systems. However, more research is needed to address the challenges and realize the full potential of these techniques.
3.2 Use Cases and Scenarios
3.2.1 Breast Cancer Classification
One of the primary applications of XAI in breast cancer recognition is in the classification of benign and malignant tumors. For instance, Jabeen et al.[9] proposed a deep learning model for breast cancer classification from ultrasound images, incorporating XAI techniques to provide interpretable visualizations of the model’s decision-making process. Similarly, Rajpal et al.[95] developed an XAI-based approach for breast cancer subtype classification using methylation data, providing insights into the biomarkers used by the model for classification. XAI has also been utilized in the classification of breast lesions. In a study by Hussain et al.[39], shape-based breast lesion classification using digital tomosynthesis images was performed. The role of XAI was significant in this study as it provided a clear understanding of the model’s decision-making process, thereby increasing the trust in AI-based diagnostic tools.
Further we discuss a simplified example of how a XAI method might be used in a real-life scenario of breast cancer classification (Algorithm 1). This example uses a XAI method, called ExplainableModel. This model is trained on a dataset of breast cancer images and associated labels (benign or malignant), and then used to predict the class of a new, unseen image. The model also provides an explanation for its prediction.
Algorithm 1. Breast Cancer Classification using XAI
Require: Training dataset D={(xi, yi)}ni=1, where xi is a breast cancer image and yi is the associated label (benign or malignant).
Require: New, unseen breast cancer image xnew.
Ensure: Predicted class ypred and explanation e for the prediction.
1: Initialize the explainable model: model←ExplainableModel()
2: Train the model on the dataset D: model.train(D)
3: Use the trained model to predict the class of xnew: ypred←model.predict(xnew)
4: Generate an explanation for the prediction: e←model.explain(xnew)
5: return ypred, e
3.2.2 Breast Cancer Detection
XAI techniques have also been applied in the detection of breast cancer from various types of medical images. For example, Rajinikanth et al.[11] used XAI to interpret the decision-making process of a model for breast cancer detection from thermal images. In another study, Prodan et al.[16] applied deep learning methods for mammography analysis and breast cancer detection, using XAI to provide interpretable explanations of the model’s predictions.
Further we discuss a simplified example of how a XAI method might be used in a real-life scenario of breast cancer classification (Algorithm 2). This example uses a XAI method, called ExplainableModel. This model is trained on a dataset of mammogram images and associated labels (cancer or no cancer), and then used to predict the presence of cancer in a new, unseen mammogram. The model also provides an explanation for its prediction.
Algorithm 2. Breast Cancer Detection using XAI
Require: Training dataset D={(xi, yi)}ni=1, where xi is a mammogram image and yi is the associated label (cancer or no cancer).
Require: New, unseen mammogram image xnew.
Ensure: Predicted label ypred and explanation e for the prediction.
1: Initialize the explainable model: model←ExplainableModel()
2: Train the model on the dataset D: model.train(D)
3: Use the trained model to predict the label of xnew: ypred←model.predict(xnew)
4: Generate an explanation for the prediction: e←model.explain(xnew)
5: return ypred, e
3.2.3 Breast Cancer Segmentation
In the task of breast cancer segmentation, XAI can provide insights into the regions that the model considers to be part of the tumor. Kadry et al.[19] used XAI to interpret the decision-making process of a model for tumor extraction in breast MRI, providing valuable insights into the model’s segmentation process.
Further we discuss a simplified example of how a XAI method might be used in a real-life scenario of breast cancer segmentation (Algorithm 3). This example uses a XAI method, which we call ExplainableModel. This model is trained on a dataset of mammogram images and associated segmentation masks (which indicate the location of cancerous tissue), and then used to predict the segmentation mask for a new, unseen mammogram. The model also provides an explanation for its prediction.
3.2.4 Breast Cancer Prognosis
XAI techniques have also been applied in predicting the prognosis of breast cancer pa-tients. For instance, Massafra et al.[96] used XAI to interpret the decision-making pro-cess of a model for predicting invasive disease events in breast cancer patients, providing insights into the factors considered by the model in its predictions.
Further we discuss a simplified example of how an XAI method might be used in a real-life scenario of breast cancer prognosis (Algorithm 4). This example uses a hypothetical XAI method, which we’ll call ExplainableModel. This model is trained on a dataset of Algorithm 3.
Algorithm 3. Breast Cancer Segmentation using XAI
Require: Training dataset D={(xi, yi)}ni=1, where xi is a patient record and yi is the associated prognostic outcome.
Require: New, unseen mammogram image xnew.
Ensure: Predicted segmentation mask mpred and explanation e for the prediction.
1: Initialize the explainable model: model←ExplainableModel()
2: Train the model on the dataset D: model.train(D)
3: Use the trained model to predict the segmentation mask for xnew: mpred←model.predict(xnew)
4: Generate an explanation for the prediction: e←model.explain(xnew)
5: return mpred, e
Patient records and associated prognostic outcomes (e.g., survival time), and then used to predict the prognosis for a new, unseen patient record. The model also provides an explanation for its prediction.
Algorithm 4. Breast Cancer Prognosis using XAI
Require: Training dataset D={(xi, yi)}ni=1, where xi is a patient record and yi is the associated prognostic outcome.
Require: New, unseen patient record xnew.
Ensure: Predicted prognostic outcome ypred and explanation e for the prediction.
1: Initialize the explainable model: model←ExplainableModel()
2: Train the model on the dataset D: model.train(D)
3: Use the trained model to predict the prognostic outcome for xnew: ypred←model.predict(xnew)
4: Generate an explanation for the prediction: e←model.explain(xnew)
5: return ypred, e
3.2.5 Breast Cancer Biomarker Discovery
XAI has also been instrumental in the discovery of biomarkers for breast cancer. In a study by Rajpal et al.[97], an XAI approach was used for biomarker discovery for breast cancer subtype classification using methylation data. The use of XAI in this context provided a clear understanding of the biomarkers used by the model for classification, thereby enhancing the interpretability of the model.
Further we discuss a simplified example of how an XAI method might be used in a real-life scenario of breast cancer biomarker discovery (Algorithm 5). This example uses a hypothetical XAI method, which we call ExplainableModel. This model is trained on a dataset of patient genomic data and associated cancer outcomes, and then used to identify potential biomarkers in a new, unseen genomic dataset. The model also provides an explanation for its findings.
3.2.6 Summary
The applications of XAI in breast cancer recognition tasks (summarized in Table 3) not only enhances the interpretability of DeepSHAP but also builds trust in their predictions. By providing visual explanations for their decisions, these models become.
Algorithm 5. Breast Cancer Biomarker Discovery using XAI
Require: Training dataset D={(xi, yi)}ni=1, where xi is a patient’s genomic data and yi is the associated cancer outcome.
Require: New, unseen genomic dataset xnew.
Ensure: Predicted potential biomarkers bpred and explanation e for the prediction.
1: Initialize the explainable model: model-ExplainableModel()
2: Train the model on the dataset D: model.train(D)
3: Use the trained model to identify potential biomarkers in xnew: bpred←model.predict(xnew)
4: Generate an explanation for the prediction: e←model.explain(xnew)
5: return bpred, e
Less of a “black box” and their predictions can be validated against the expert knowledge of clinicians. This is particularly important in the medical field, where the stakes are high and the predictions of these models can have significant implications for patient care.
Table 3. Applications of XAI in Breast Cancer Recognition
Application Area |
Technique |
Description |
Breast cancer classification |
Deep learning and XAI |
Jabeen et al.[9]: Classification of tumors using ultrasound images and interpretable visualizations. Rajpal et al.[95]: Subtype classification using methylation data with insights into biomarkers. |
Hussain et al.[39]: Shape-based lesion classification with digital tomosynthesis images. |
||
Breast cancer detection |
Deep learning and XAI |
Rajinikanth et al.[11]: Detection from thermal images with in- terpretable decision-making. Prodan et al.[16]: Mammography analysis for cancer detection with model prediction explanations. |
Breast cancer segmentation |
Tumor segmentation |
Kadry et al.[19]: Interpretation of MRI-based tumor extraction. |
Breast cancer prognosis |
Prognostic prediction |
Massafra et al.[96]: Prediction of invasive disease events with insights into decision factors. |
Breast cancer biomarker discovery |
Biomarker identification |
Rajpal et al.[97]: Biomarker discovery using methylation data with clear understanding of utilized biomarkers. |
3.3 Evaluation of Explanations
The evaluation of explanations generated by XAI models is crucial to ensure their effectiveness and reliability. The need for quantitative evaluation metrics arises from the necessity to objectively assess the quality of explanations and compare different XAI methods. These metrics provide a standardized measure of evaluation, enabling a fair comparison across different models and domains. They can assess various aspects of explanations, such as their fidelity to the original model, their interpretability, or their usefulness to the end-user[98,99].
Several approaches have been proposed to evaluate the quality of explanations provided by XAI models. These approaches can be broadly categorized into two types: user-centric evaluations and model-centric evaluations.
(1) User-centric evaluations focus on the usefulness of explanations to the end-user. They often involve user studies where human subjects interact with the XAI system and provide feedback on the quality of explanations. These evaluations can assess various aspects of explanations, such as their understandability, usefulness in decision-making, and their impact on user trust in the AI system[100].
(2) Model-centric evaluations, on the other hand, focus on the fidelity of explanations to the original model. They often involve comparing the predictions of the original model with those of the explanation model. High fidelity indicates that the ex-planation model accurately represents the decision-making process of the original model.
XAI methods provide clarity on how AI models derive their predictions, which is essential in clinical settings for validating and trusting AI-assisted diagnoses. Different XAI models return different types of explanatory information, primarily focusing on the features and reasoning that underpin their decision-making processes. Here, we introduce the primary forms of explanations provided by key XAI methods employed in breast cancer diagnostics.
LIME explains predictions by approximating the local decision boundary of any classifier with an interpretable model. It highlights which features in a specific instance (e.g., a mammogram or histopathological image) most influence the model’s prediction. For instance, LIME might indicate that the presence of irregular mass shapes or specific texture patterns strongly suggests malignancy in breast cancer diagnosis.
SHAP values explain the output of any model by computing the contribution of each feature to the prediction. These values are based on game theory and provide a fair distribution of the prediction output among the features. In breast cancer, SHAP can elucidate which clinical parameters (like tumor size, age of the patient, or genetic markers) and image features (such as lesion density or margin characteristics) are most impactful in models predicting cancer stages or treatment responses.
Grad-CAM uses the gradients of any target concept (say, “malignant” or “benign”) flowing into the final convolutional layer to produce a coarse localization map highlighting the important regions in the image for predicting the concept. For breast cancer, Grad-CAM can visually demonstrate areas in an image critical for the model’s decision, such as highlighting regions of a tumor suspected to have higher malignancy potential.
DeepLIFT compares the activation of each neuron to its “reference activation” and assigns contribution scores according to the difference caused by each feature. In breast cancer diagnostics, DeepLIFT can identify not just the features (e.g., edges of a lesion in mammography) but also specific characteristics of the patient’s genomic profile that influence the predictive model.
The interpretability facilitated by these methods not only aids clinicians in understanding the AI’s reasoning but also assists in validating the AI’s reliability and accuracy in clinical applications. This transparency is critical for integrating AI tools into routine clinical practice, ensuring that they complement traditional diagnostic techniques and contribute to more accurate and personalized patient care. However, it is important to note that the choice of evaluation approach and metrics should be guided by the specific needs and constraints of the application domain. For instance, in safety-critical domains such as healthcare, the fidelity of explanations might be prioritized over their interpretability to ensure that the explanations accurately represent the decision-making process of the AI model[101].
Despite the progress made in developing evaluation metrics for XAI, there are still challenges that need to be addressed. For instance, there is a lack of quantitative evaluation metrics for some properties of explanations, such as clarity, and for some types of explanations, such as example-based methods[98]. The evaluation of XAI models often involves a trade-off between different properties of explanations, such as interpretability and fidelity. Future research should aim to develop evaluation metrics that can balance these trade-offs and provide a comprehensive assessment of explanation quality.
3.4 Integration into Clinical Workflow
The integration of XAI into the clinical workflow is a complex process that requires careful consideration of various factors. The successful adoption of XAI in clinical settings hinges on addressing these challenges and developing strategies for seamless integration and user-friendly interfaces. One of the primary obstacles is the need for high computational power and large datasets for training and validating AI models. The management, analysis, and interpretation of big data in healthcare can be a daunting task, and healthcare providers need to be equipped with the appropriate infrastructure to handle this data effectively[102]. The integration of AI into clinical workflows also requires careful consideration of ethical and regulatory issues. The use of patient data in AI models raises concerns about patient privacy and data security. Regulatory bodies also need to establish guidelines for the use of AI in healthcare to ensure that these technologies are used responsibly and ethically[103]. Despite these challenges, there are several strategies that can facilitate the integration of XAI into the clinical workflow. One approach is to develop user-friendly interfaces that allow clinicians to interact with AI models easily. These interfaces should be designed to present AI-generated insights in a clear and understandable manner, enabling clinicians to make informed decisions based on these insights.
A process used for AI-based breast cancer detection and emphasizes continuous improvement through feedback loops and decision points was delineated in Figure 1. This detailed portrayal underscores the application of advanced technologies and methodologies to enhance the accuracy and reliability of breast cancer diagnostics. The process initiates with the collection of critical diagnostic data such as mammograms, ultrasounds, and MRIs. This step is essential as it gathers the raw images and data required to detect and diagnose breast cancer using AI. Following data collection, the next phase involves preprocessing, which typically includes normalization to standardize the data and augmentation to artificially expand the dataset. These steps are crucial for preparing the data for effective training of AI models by enhancing the dataset’s diversity and reducing model overfitting. After preprocessing, the data is used to train deep learning algorithms. This training process involves feeding the data through complex neural network architectures that learn to identify patterns and features indicative of breast cancer. The effectiveness of this step hinges on the quality and variety of the data provided. Post-training, the model undergoes a validation phase where its performance is rigorously tested against a set of clinical data that was not part of the training dataset. This step is crucial to ensure that the model performs well in real-world scenarios and can generalize beyond the training examples. Validated models are then applied to make predictions regarding breast cancer presence in new, unseen medical images. This step marks the practical use of the trained AI in clinical environments, providing preliminary diagnoses based on learned patterns. To enhance the trustworthiness and transparency of AI decisions, XAI methods such as LIME and SHAP are used to generate explanations for the AI’s predictions. These explanations help clinicians understand why the model made a particular decision, highlighting the features or factors that most influenced the outcome. Based on the AI’s predictions and the accompanying explanations, a diagnostic decision is made to determine the presence of breast cancer. This decision supports clinical diagnosis and can significantly impact the subsequent treatment plan.
Figure 1. Process of AI-based Breast Cancer Detection.
The process includes several feedback mechanisms: (1) Post-diagnosis, the outcomes and decisions are reviewed by clinicians. This feedback is crucial for assessing the practical effectiveness and clinical relevance of the AI predictions. (2) Both clinical feedback and ongoing performance assessments feed back into the model training phase. This continuous improvement loop allows for refinements and adjustments to the AI algorithms based on real-world outcomes and evolving medical knowledge.
At several stages, conditional checks are performed: (1) After validation, the model’s accuracy is assessed to determine if it meets the required thresholds for clinical deployment. (2) The confidence level of the AI’s predictions is evaluated. If the confidence is high, the process moves forward; if not, adjustments may be needed. (3) Finally, the AI’s decisions are compared against clinical expectations. If there is a match, the process continues smoothly; discrepancies might trigger a reevaluation or further refinement of the model.
This comprehensive process not only ensures the technical efficacy of AI models in diagnosing breast cancer but also integrates clinical insights and validations, making AI a valuable partner in the fight against breast cancer.
4 CHALLENGES AND FUTURE DIRECTIONS
The application of XAI in breast cancer recognition tasks has shown promising results, but there are still several challenges to overcome and future directions to explore.
One of the main challenges in XAI is the trade-off between model interpretability and performance. While simpler models are often more interpretable, they may not perform as well as more complex models, which can be harder to interpret. This trade-off is particularly relevant in healthcare, where high performance is crucial for patient outcomes, but interpretability is also important for gaining the trust of clinicians and patients[104]. Another challenge is the lack of standardized evaluation metrics for explanations. While several metrics have been proposed, there is no consensus on which metrics are the most appropriate for evaluating the quality of explanations. This makes it difficult to compare different XAI methods and to assess their effectiveness[105]. The integration of XAI into clinical workflows poses its own set of challenges. These include technical challenges, such as the need for high computational power and large datasets, as well as ethical and regulatory challenges related to patient privacy and data security[102,103]. Despite these challenges, there are several promising directions for future research in XAI for breast cancer recognition. One direction is the development of new methods that can provide high-quality explanations without sacrificing model performance. This could involve the use of hybrid models that combine the strengths of different types of models, or the development of new techniques for generating explanations[104]. Another direction is the development of standardized evaluation metrics for explanations. This could involve the establishment of benchmarks for different types of explanations, or the development of new metrics that can capture the quality of explanations more accurately[105]. Finally, there is a need for more research on the integration of XAI into clinical workflows. This could involve the development of user-friendly interfaces for interacting with AI models, or the exploration of strategies for integrating AI into existing clinical workflows in a way that complements rather than replaces the work of clinicians[102,103].
5 CONCLUSION
This mini-review has explored the applications of XAI in various breast cancer recognition tasks, such as classification and segmentation. We have discussed the importance of XAI in enhancing the interpretability of DeepSHAP, which is crucial for gaining the trust of clinicians and patients. We have also highlighted the challenges in adopting XAI methods in clinical settings, including the trade-off between model interpretability and performance, the lack of standardized evaluation metrics for explanations, and the technical, ethical, and regulatory challenges related to the integration of XAI into clinical workflows. Despite these challenges, we have identified several promising directions for future research, including the development of new XAI methods, the standardization of evaluation metrics, and the integration of XAI into clinical workflows.
The application of XAI methods in breast cancer recognition has the potential to significantly impact the management of breast cancer. By providing interpretable explanations for their predictions, XAI methods can help clinicians to make more informed decisions about diagnosis and treatment. This can lead to improved patient outcomes, as well as increased trust in AI technologies among clinicians and patients. The integration of XAI into clinical workflows can improve the efficiency of clinical processes, freeing up clinicians to focus on more complex aspects of patient care.
Despite the progress that has been made in the field of XAI, there is still much work to be done. Continued research and development in XAI techniques is crucial for addressing the existing challenges and realizing the full potential of XAI in breast cancer recognition. This includes the development of new XAI methods that can provide high-quality explanations without sacrificing model performance, the establishment of standardized evaluation metrics for explanations, and the exploration of strategies for integrating XAI into clinical workflows. By advancing our understanding and capabilities in XAI, we can pave the way for more effective and trustworthy AI applications in breast cancer recognition and beyond.
The XAI methods have shown promise in enhancing the interpretability and trustworthiness of AI models in breast cancer recognition. By providing insights into the decision-making process of these models, XAI methods can empower clinicians to make more informed decisions and improve patient outcomes. The continued development and refinement of XAI techniques have the potential to revolutionize the way AI-based diagnostic tools are employed in breast cancer management. Further research should focus on the development of domain-specific XAI techniques, the evaluation of explanation quality, and the seamless integration of XAI methods into the clinical workflow to maximize their impact on patient care.
Acknowledgements
Not applicable.
Conflicts of Interest
The author declared no conflict of interest.
Data Availability
No additional data are available.
Copyright Permissions
Copyright © 2024 The Author(s). Published by Innovation Forever Publishing Group Limited. This open-access article is licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, sharing, adaptation, distribution, and reproduction in any medium, provided the original work is properly cited.
Author Contribution
Damaševičius R was responsible for the collection, compilation and summary of all data for articles.
Abbreviation List
AI, Artificial intelligence
BRCA, Breast invasive carcinoma
CNNs, Convolutional neural networks
DBT, Digital breast tomosynthesis
DeepSHAP, Deep shapley additive explanations
Grad-CAM, Gradient-weighted class activation mapping
LIME, Local interpretable model-agnostic explanations
MRI, Magnetic resonance imaging
SHAP, Shapley additive explanations
XAI, Explainable AI
References
[1] Nazir S, Dickson D, Akram M. Survey of explainable artificial intelligence techniques for biomedical imaging with deep neural networks. Comput Biol Med, 2023; 156: 106668.[DOI]
[2] Le E, Wang Y, Huang Y et al. Artificial intelligence in breast imaging. Clin Radiol, 2019; 74: 357-366.[DOI]
[3] Binder A, Bockmayr M, Hägele M et al. Morphological and molecular breast cancer profiling through explainable machine learning. Nat Mac Intell, 2021; 3: 355-366.[DOI]
[4] Silva-Aravena F, Núñez Delafuente H, Gutiérrez-Bahamondes JH et al. A Hybrid Algorithm of ML and XAI to Prevent Breast Cancer: A Strategy to Support Decision Making. Cancers, 2023; 15: 2443.[DOI]
[5] Altini N, Puro E, Taccogna M et al. Tumor Cellularity Assessment of Breast Histopathological Slides via Instance Segmentation and Pathomic Features Explain-ability. Bioengineering, 2023; 10: 396.[DOI]
[6] Zebari D, Ibrahim D, Zeebaree D et al. Systematic Review of Computing Approaches for Breast Cancer Detection Based Computer Aided Diagnosis Using Mammogram Images. Appl Artif Intell, 2021; 35: 2157-2203.[DOI]
[7] Kumar S, Das A. Peripheral blood mononuclear cell derived biomarker detection using eXplainable Artificial Intelligence (XAI) provides better diagnosis of breast cancer. Comp Bio Chem, 2023; 104: 107867.[DOI]
[8] Kumar Vijay, Singh Amit Kumar, Damasevicius Robertas. Guest Editorial Ar-tificial Intelligence-Driven Biomedical Imaging Systems for Precision Diagnostic. Applications IEEE. J Bio Heal Inform, 2024; 28: 1158-1160.[DOI]
[9] Jabeen K, Khan M, Alhaisoni M et al. Breast Cancer Classification from Ultrasound Images Using Probability-Based Optimal Deep Learning Feature Fusion. Sensors, 2022; 22: 807.[DOI]
[10] Irfan R, Almazroi A, Rauf H et al. Dilated semantic segmentation for breast ultrasonic lesion detection using parallel feature fusion. Diagnostics, 2021; 11: 1212.[DOI]
[11] Rajinikanth V, Kadry S, Taniar D et al. Breast-Cancer Detection using Thermal Images with Marine-Predators-Algorithm Selected Features: Proceedings of 2021 IEEE 7th International Conference on Bio Signals, Images and Instrumentation. Chennai, India, 25-27 March 2021.[DOI]
[12] Jin D, Sergeeva E, Weng W et al. Explainable deep learning in healthcare: A methodological survey from an attribution view. WIREs Mech Dis, 2022; 14: e1548.[DOI]
[13] Zhang Y, Weng Y, Lund J. Applications of Explainable Artificial Intelligence in Diagnosis and Surgery. Diagnostics, 2022; 12: 237.[DOI]
[14] Gu D, Zhao W, Xie Y et al. A personalized medical decision support system based on explainable machine learning algorithms and ecc features: Data from the real world. Diagnostics, 2021; 11: 1677.[DOI]
[15] Scapicchio C, Lizzi F, Fantacci M. Explainability of a CNN for breast density assessment. Il Nuovo Cimento C, 2021; 44: 1-4.[DOI]
[16] Prodan M, Paraschiv E, Stanciu A. Applying Deep Learning Methods for Mammography Analysis and Breast Cancer Detection. Appl Sci, 2023; 13: 4272.[DOI]
[17] Meraj T, Alosaimi W, Alouffi B et al. A quantization assisted U-Net study with ICA and deep features fusion for breast cancer identification using ultrasonic data. PeerJ Comp Sci, 2021; 7: e805.[DOI]
[18] Rasaee H, Rivaz H. Explainable AI and susceptibility to adversarial attacks: A case study in classification of breast ultrasound images. 2021 IEEE International Ultrasonics Symposium, 2021: 1-4.[DOI]
[19] Kadry S, Damasevicius R, Taniar D et al. Extraction of Tumour in Breast MRI using Joint Thresholding and Segmentation-A Study. 2021 Seventh International conference on Bio Signals, Images, and Instrumentation (ICBSII), 2021: 1-5.[DOI]
[20] Kaur Amandeep, Kaushal Chetna, Sandhu Jasjeet Kaur et al. Histopathological Image Diagnosis for Breast Cancer Diagnosis Based on Deep Mutual Learning. Diagnostics, 2024; 14: 95.[DOI]
[21] Jung J, Lee H, Jung H et al. Essential properties and explanation effectiveness of explainable artificial intelligence in healthcare: A systematic review. Heliyon, 2023; 9: e16110.[DOI]
[22] Gulum M, Trombley C, Kantardzic M. A review of explainable deep learning cancer detection models in medical imaging. Appl Sci, 2021; 11 4573.[DOI]
[23] Loizidou K, Elia R, Pitris C. Computer-aided breast cancer detection and classification in mammography: A comprehensive review. Comp Biolo Med, 2023; 153: 106554.[DOI]
[24] Yang W, Dempsey P. Diagnostic Breast Ultrasound: Current Status and Future Directions. Radiolo Clin N Am, 2007; 45: 845-861.[DOI]
[25] Brunetti N, Calabrese M, Martinoli C et al. Artificial Intelligence in Breast Ultrasound: From Diagnosis to Prognosis-A Rapid Review. Diagnostics, 2023; 13: 58.[DOI]
[26] Gao Y, Reig B, Heacock L et al. Magnetic Reso-nance Imaging in Screening of Breast Cancer. Radiolo Clin N Am, 2021; 59: 85-98.[DOI]
[27] White M, Soman A, Weinberg C et al. Factors associated with breast MRI use among women with a family history of breast cancer. Breast J. 2018; 24: 764-771.[DOI]
[28] Chong A, Weinstein S, McDonald E et al. Digital breast tomosyn-thesis: Concepts and clinical practice. Radiology, 2019; 292: 1-14.[DOI]
[29] Sechopoulos I, Teuwen J, Mann R. Artificial intelligence for breast cancer detec-tion in mammography and digital breast tomosynthesis: State of the art. Semin Cancer Biol, 2021; 72: 214-225.[DOI]
[30] Bai J, Posner R, Wang T et al. Applying deep learning in digital breast tomosynthesis for automatic breast cancer detection: A review. Med Image Anal, 2021; 71: 102049.[DOI]
[31] Nassif A, Talib M, Nasir Q et al. Breast cancer detec-tion using artificial intelligence techniques: A systematic literature review. Artif Intell Med, 2022; 127: 102276.[DOI]
[32] Baughan N, Douglas L, Giger M. Past, Present, and Future of Machine Learn-ing and Artificial Intelligence for Breast Cancer Screening. J Breast Imaging, 2022; 4: 451-459.[DOI]
[33] Houssami N, Kirkpatrick-Jones G, Noguchi N et al. Artificial Intelligence (AI) for the early detection of breast cancer: a scoping review to assess AI’s poten-tial in breast screening practice. Expert Rev Med Devic, 2019; 16: 351-362.[DOI]
[34] Uzun Ozsahin D, Ikechukwu Emegano D, Uzun B et al. The Systematic Re-view of Artificial Intelligence Applications in Breast Cancer Diagnosis. Diagnostics, 2023; 13: 45.[DOI]
[35] Shah S, Khan R, Arif S et al. Artificial intelligence for breast cancer analysis: Trends & directions. Comp Biol Med, 2022; 142: 105221.[DOI]
[36] Sadoughi F, Kazemy Z, Hamedan F et al. Artificial intelligence methods for the diagnosis of breast cancer by image processing: A review. Breast Cancer, 2018; 10: 219-230.[DOI]
[37] Maqsood S, Damasevicius R, Maskeliunas R. TTCNN: A Breast Cancer Detection and Classification towards Computer-Aided Diagnosis Using Digital Mammography in Early Stages. Appl Sci, 2022; 12: 3273.[DOI]
[38] Zebari D, Ibrahim D, Zeebaree D et al. Breast cancer detection using mammogram images with improved multi-fractal dimension approach and feature fusion. Appl Sci, 2021; 11: 12122.[DOI]
[39] Hussain S, Buongiorno D, Altini N et al. Shape-Based Breast Lesion Clas-sification Using Digital Tomosynthesis Images: The Role of Explainable Artificial Intelligence. Appl Sci, 2022; 12: 6230.[DOI]
[40] Barzaman K, Karami J, Zarei Z et al. Breast cancer: Biology, biomarkers, and treatments. Int Immunopharmacol, 2020; 84: 106535.[DOI]
[41] Ross J, Gay L. Comprehensive genomic sequencing and the molecular profiles of clinically advanced breast cancer. Pathol, 2017; 49: 120-132.[DOI]
[42] Weigelt B, Pusztai L, Ashworth A et al. Challenges translating breast cancer gene signatures into the clinic. Nat Rev Clinl Oncol, 2012; 9: 58-64.[DOI]
[43] Atehortua N, Issa A. A method to measure clinical practice patterns of breast cancer genomic diagnostics in health systems. Pers Med, 2012; 9: 585-592.[DOI]
[44] Marcom P, Barry W, Datto M et al. A randomized phase II trial evaluating the performance of genomic expression profiles to direct the use of preoperative chemotherapy for early-stage breast cancer. J Clin Oncol, 2010; 28: 15.[DOI]
[45] Kwa M, Makris A, Esteva F. Clinical utility of gene-expression signatures in early stage breast cancer. Nat Rev Clin Oncol, 2017; 14: 595-610.[DOI]
[46] Low S, Zembutsu H, Nakamura Y. Breast cancer: The translation of big genomic data to cancer precision medicine. Cancer Sci, 2018; 109: 497-506.[DOI]
[47] Huang S, Yang J, Fong S et al. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett, 2020; 471: 61-71.[DOI]
[48] Sheth D, Giger M. Artificial intelligence in the interpretation of breast cancer on MRI. J Magn Reson Imag, 2020; 51: 1310-1324.[DOI]
[49] Adadi A, Berrada M. Peeking Inside the Black-Box: A Survey on Explainable Artificial Intelligence (XAI). IEEE Access, 2018; 6: 52138-52160.[DOI]
[50] Velden B, Kuijf H, Gilhuijs K et al. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med Imag Anal, 2022; 79: 102479.[DOI]
[51] Qian J, Li H, Wang J et al. Recent Advances in Explainable Artificial Intelligence for Magnetic Resonance Imaging. Diagnostics, 2023; 13: 1571.[DOI]
[52] Lamy J, Sekar B, Guezennec G et al. Explainable artificial intelligence for breast cancer: A visual case-based reasoning approach. Artif Intel Med, 2019; 94: 42-53.[DOI]
[53] Ness L, Barkan E, Ozery-Flato M. Improving the Performance and Explainability of Mammogram Classifiers with Local Annotations: Proceedings of the Interpretable and Annotation-Efficient Learning for Medical Image Computing. Lima, Peru, 2 October 2020.[DOI]
[54] Saranya A, Subhashini R. A systematic review of Explainable Artificial Intelli-gence models and applications: Recent developments and future trends. Decis Anal J, 2023; 7: 100230.[DOI]
[55] Saeed W, Omlin C. Explainable AI (XAI): A systematic meta-survey of current challenges and future opportunities. Knowl-Based Syst, 2023; 263: 110273.[DOI]
[56] Ribeiro Marco Tulio, Singh Sameer, Guestrin Carlos. “Why should I trust you?” Explaining the predictions of any classifier: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. California, USA, 13 August 2016.[DOI]
[57] Lundberg Scott M, Lee Su-In. A Unified Approach to Interpreting Model Predictions: Proceedings of the 31st International Conference on Neural Information Processing Systems. California, USA, 4-9 December 2017.[DOI]
[58] Zhang Q. Prognostic Model and Influencing Factors for Breast Cancer Patients. Int J Biol Life Sci, 2023; 2: 209-219.[DOI]
[59] Zhao X, Jiang C. The prediction of distant metastasis risk for male breast cancer patients based on an interpretable machine learning model. BMC Med Inform Decis Mak, 2023; 74: 23.[DOI]
[60] Çubuk C, Loucera C, Peña-Chilet M et al. Crosstalk between Metabo-lite Production and Signaling Activity in Breast Cancer. Int J Mol Sci, 2023; 24: 7450.[DOI]
[61] Mendonca-Neto R, Reis J, Okimoto L et al. Classification of breast cancer subtypes: A study based on representative genes. J Brazil Com Soc, 2022; 28: 59-68.[DOI]
[62] Selvaraju R, Cogswell M, Das A et al. Grad-CAM: Visual Explanations from Deep Net-works via Gradient-Based Localization. Int J Comput Vision, 2019; 128: 336-359.[DOI]
[63] Hakkoum H, Idri A, Abnane I. Assessing and Comparing Interpretability Tech-niques for Artificial Neural Networks Breast Cancer Classification. Comput Methods Biomech Biomed Eng Imaging Vis, 2021; 9: 587-599.[DOI]
[64] Almutairi S, Manimurugan S, Kim B et al. Breast cancer classification using Deep Q Learning (DQL) and gorilla troops optimization (GTO). Appl Soft Comput, 2023; 142: 110292.[DOI]
[65] Modhukur V, Sharma S, Mondal M et al. Machine Learning Approaches to Classify Primary and Metastatic Cancers Using Tissue of Origin-Based DNA Methylation Profiles. Cancers, 2021; 13: 3768.[DOI]
[66] Meshoul S, Batouche A, Shaiba H et al. Explainable Multi-Class Classifi-cation Based on Integrative Feature Selection for Breast Cancer Subtyping. Mathematics, 2022; 10: 4271.[DOI]
[67] Nahid A, Raihan M, Bulbul A. Breast cancer classification along with feature prioritization using machine learning algorithms. Health Technol (Berl), 2022; 12: 1061-1069.
[68] Hossain A, Nisha J, Johora F. Breast Cancer Classification from Ul-trasound Images using VGG16 Model based Transfer Learning. Int J Imag, 2023; 15: 12-22.[DOI]
[69] Lou Q, Li Y, Qian Y et al. Mammogram classification based on a novel convolutional neural network with efficient channel attention. Comput Biol Med, 2022; 150: 106802.[DOI]
[70] Masud M, Eldin R, Hossain M. Convolutional neural network-based models for diagnosis of breast cancer. Neural Comput Appl, 2022; 34: 11383-11394.[DOI]
[71] Ricciardi R, Mettivier G, Staffa M et al. A deep learning classifier for digital breast tomosynthesis. Phys Med, 2021; 83: 184-193.[DOI]
[72] Eskandari A, Du H, AlZoubi A. Towards Linking CNN Decisions with Cancer Signs for Breast Lesion Classification from Ultrasound Images. In: Papież BW, Yaqub M, Jiao J, Namburete AI, Noble JA (eds). Medical Image Understanding and Analysis. MIUA 2021. Lecture Notes in Computer Science, Springer, Cham.2021; 12722: 423-437.[DOI]
[73] Lee Y, Huang C, Shih C et al. Axillary lymph node metastasis status pre-diction of early-stage breast cancer using convolutional neural networks. Comput Biol Med, 2021; 130: 104206.[DOI]
[74] Hakkoum H, Idri A, Abnane I. Artificial Neural Networks Interpretation Using LIME for Breast Cancer Diagnosis. In: Rocha Á, Adeli H, Reis L, Costanzo S, Orovic I, Moreira F (eds). Trends and Innovations in Information Systems and Technologies. WorldCIST 2020. Advances in Intelligent Systems and Computing, Springer, Cham., 2020; 1161: 15-24.[DOI]
[75] Mathew T. An Optimized Extremely Randomized Tree Model For Breast Cancer Classification. J Theor Appl Inform Technol, 2022; 100: 5234-5246.
[76] Keren E, Angeline K, Glory P. Prediction of Breast Cancer Recurrence in Five Years using Machine Learning Techniques and SHAP. Intelligent Computing Techniques for Smart Energy Systems: Proceedings of ICTSES 2021. Singapore: Springer Nature Singapore.[DOI]
[77] Mohi U, Biswas N, Rikta S et al. XML-LightGBMDroid: A self-driven interactive mobile application utilizing explainable machine learning for breast cancer diagnosis. Eng Rep, 2023; 5: e12666.[DOI]
[78] Masud M, Hossain M, Alhumyani H et al. Pre-Trained Convolutional Neural Networks for Breast Cancer Detection Using Ultrasound Images. ACM Trans Internet Technol, 2021; 21: 1-17.[DOI]
[79] Chatterjee C, Krishna G. A novel method for IDC prediction in breast cancer histopathology images using deep residual neural networks: Proceedings of the 2019 2nd International Conference on Intelligent Communication and Computational Techniques. Jaipur, India, 28-29 September 2019.[DOI]
[80] Oya M, Sugimoto S, Sasai K et al. Investigation of clinical target vol-ume segmentation for whole breast irradiation using three-dimensional convolu-tional neural networks with gradient-weighted class activation mapping. Radiol Phys Technol, 2021; 14: 238-247.[DOI]
[81] Chaudhury S, Sau K, Khan M et al. Deep transfer learning for IDC breast cancer detection using fast AI technique and Sqeezenet architecture. Math Biosci Eng, 2023; 20: 10404-10427.[DOI]
[82] Deb S, Abhishek A, Jha R. 2-Stage Convolutional Neural Network for Breast Cancer Detection from Ultrasound Images: Proceedings of the 2023 National Conference on Communications. Guwahati, India, 23-26 February 2023.[DOI]
[83] van der Velden BHM, Ragusi MAA, Janse MHA et al. Interpretable deep learning regression for breast density estimation on MRI. Medical Imaging 2020: Computer-Aided Diagnosis, Event: SPIE Medical Imaging, 2020, Houston, Texas, United States. 2020; 1131412.[DOI]
[84] Maouche I, Terrissa L, Benmohammed K et al. An Explainable AI approach for Breast Cancer Metastasis Prediction based on Clinicopathological data. IEEE Trans Biom Eng, 2023; 70: 3321-3329.[DOI]
[85] Doan L, Angione C, Occhipinti A. Introduction: Machine Learning Methods for Survival Analysis with Clinical and Transcriptomics Data of Breast Cancer. In: Methods in Molecular Biology. Publishing: Humana Press, British, 2022; 325-393.
[86] Moncada-Torres A, Maaren M, Hendriks M et al. Explainable machine learning can outperform Cox regression predictions and provide insights in breast cancer survival. Sci Rep, 2021; 11: 6968.[DOI]
[87] Jansen T, Geleijnse G, Maaren M et al. Machine learning explainability in breast cancer survival./Digital Personalized Health and Medicine. IOS Press, 2020: 307-311.[DOI]
[88] Yu H, Chen F, Lam K et al. Potential Determinants for Radiation-Induced Lymphopenia in Patients With Breast Cancer Using Interpretable Machine Learning Approach. Front Immunol, 2022; 13: 768811.[DOI]
[89] Xu L, Guo C. CoxNAM: An interpretable deep survival analysis model. Expert Syst Appl, 2023; 227: 120218.[DOI]
[90] Cordova C, Muñoz R, Olivares R et al. HER2 classification in breast cancer cells: A new explainable machine learning application for immunohistochemistry. Oncol Let, 2023; 25: 1-9.[DOI]
[91] Massi M, Dominoni L, Ieva F et al. A Deep Survival EWAS approach estimating risk profile based on pre-diagnostic DNA methylation: An application to breast cancer time to diagnosis. PLoS Comput Biol, 2022; 18: e1009959.[DOI]
[92] Langlotz C, Allen B, Erickson B et al. A Roadmap for Foun-dational Research on Artificial Intelligence in Medical Imaging: From the 2018 NIH/RSNA/ACR/The Academy Workshop. Radiology, 2019; 291: 781-791.[DOI]
[93] Zhu J, Liapis A, Risi S et al. Explainable AI for Designers: A Human-Centered Perspective on Mixed-Initiative Co-Creation: Proceedings of the IEEE Conference on Compu-tational Intelligence and Games. Maastricht, Netherlands, 14-17 August 2018.[DOI]
[94] Leichtmann B, Humer C, Hinterreiter A et al. Effects of Explainable Artificial Intelligence on trust and human behavior in a high-risk decision task. Com Hum Behav, 2022; 139: 107539.[DOI]
[95] Rajpal S, Rajpal A, Saggar A et al. XAI-MethylMarker: Explainable AI approach for biomarker discovery for breast cancer subtype classification using methylation data. Exp Sys Appl, 2023; 225: 120130.[DOI]
[96] Massafra R, Fanizzi A, Amoroso N et al. Analyzing breast cancer invasive disease event classification through explainable artificial intelligence. Front Med, 2023; 10: 1116354.[DOI]
[97] Rajpal S, Rajpal A, Agarwal M et al. XAI-CNVMarker: Explainable AI-based copy number variant biomarker discovery for breast cancer subtypes. Biomed Signal Proces, 2023; 84: 104979.[DOI]
[98] Markus A, Kors J, Rijnbeek P. The role of explainability in creating trustworthy artificial intelligence for health care: A comprehensive survey of the terminology, design choices, and evaluation strategies. J Bio Inform, 2021; 113: 103655.[DOI]
[99] Kontham RR, Kondoju AK, Fouda MM et al. An end-to-end explainable AI system for analyzing breast cancer prediction models. 2022 IEEE International Conference on Internet of Things and Intelligence Systems, 2022: 402-407.[DOI]
[100] Larasati R. Explainable AI for breast cancer diagnosis: Application and user’s understandability perception. 2022 International Conference on Electrical, Computer and Energy Technologies, 2022: 1-6.[DOI]
[101] Islam M, Ahmed M, Barua S et al. A Systematic Review of Explainable Artificial Intelligence in Terms of Different Applica-tion Domains and Tasks. Appl Sci, 2022; 12: 1353.[DOI]
[102] Dash S, Shakyawar S, Sharma M et al. Big data in healthcare: management, analysis and future prospects. J Big Data, 2019; 6: 54.[DOI]
[103] Luo J, Wu M, Gopukumar D et al. Big Data Application in Biomedical Research and Health Care: A Literature Review. Biomed Inform Insights, 2016; 8: 1-10.[DOI]
[104] Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell, 2019; 1: 206-215.[DOI]
[105] Doshi-Velez F, Kim B. Towards a rigorous science of interpretable machine learning. arXiv, 2020; 1: 1-13.[DOI]
Brief of Corresponding Author(s)
Robertas Damaševičius He is a Professor at the Department of Applied Informatics at Vytautas Magnus University. He holds a PhD in Informatics Engineering from Kaunas University of Technology, where he also completed his MSc and BSc in Informatics. His academic career is marked by a strong focus on software engineering, artificial intelligence, and computer science. With over 500 publications in international journals and conferences, he has established a significant presence in the research community. His work has earned him an h-index of 73. |

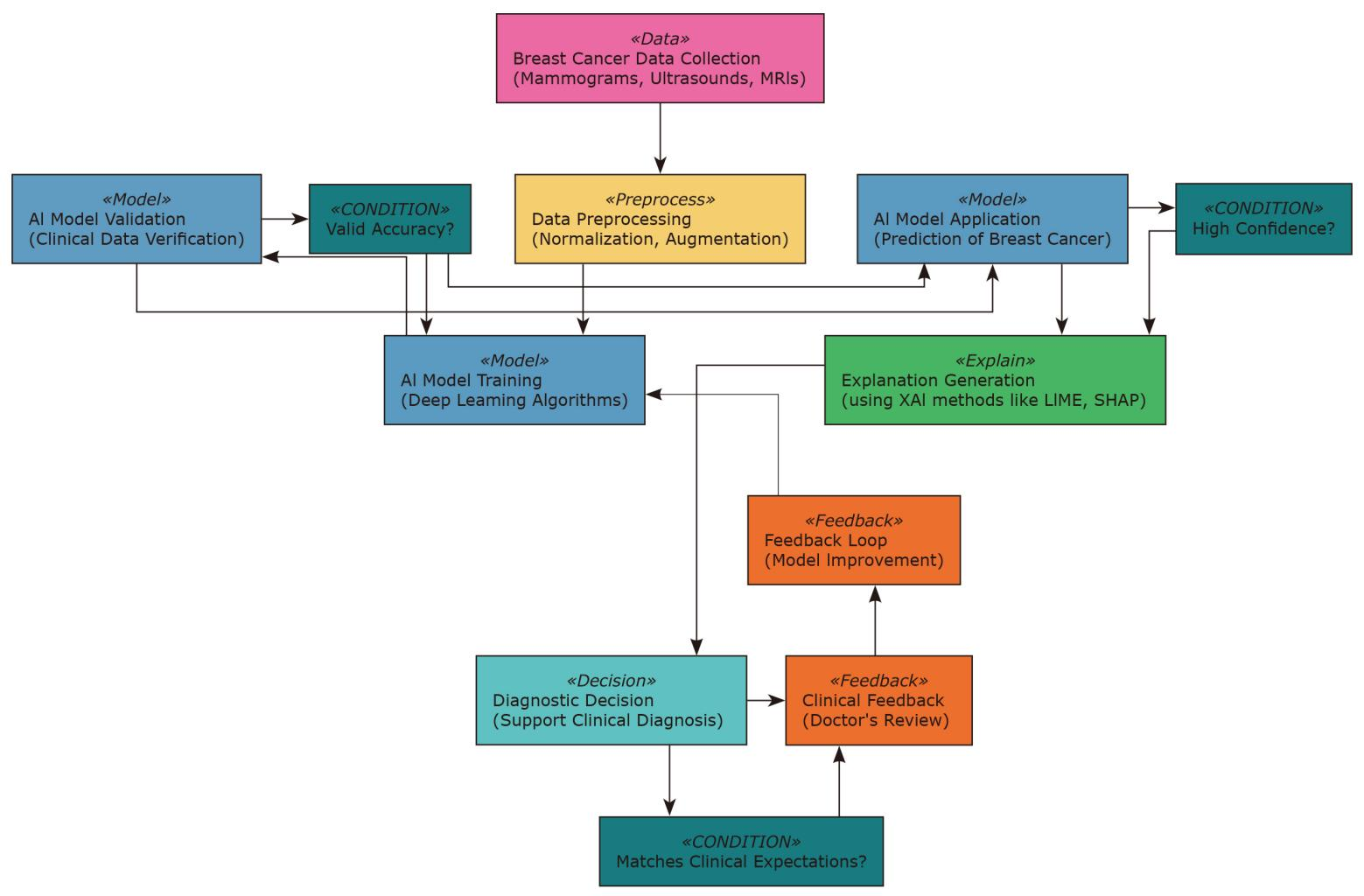




 Copyright ©
Copyright ©